Integrated approaches for the discovery of novel enzymatic activities
•
0 likes•195 views
The document summarizes the work of the "Novel Enzymatic Activities" group at LABGeM in discovering new enzymatic activities. The group uses several bioinformatics methods like CanOE, ASMC, and reaction molecular signature networks to integrate genomic and structural data to classify enzyme families and predict orphan enzymes. The group has 3 researchers, 2 PhD students, and 1 undergraduate student working on projects like elucidating the function of unknown protein families and exploring archaeal enzyme activities to reduce the number of orphan enzymes and propose new enzymatic functions.
Report
Share
Report
Share
Download to read offline
Recommended
CECAM-2015_ESR10_poster_Andrew-Brockman

This research aims to identify protein-protein interactions in Anopheles gambiae that could be targeted by ligands to block transmission of malaria. Computational methods were used to predict nearly 10,000 putative interactions and 100 interaction modules. The interactions were identified using various data sources and techniques including gene expression, regulatory motifs, orthology, and literature mining. Validation with additional methods like structural analysis could help confirm some of the predicted interactions. The goal is to develop novel drugs, insecticides or repellents by disrupting interactions essential for malaria transmission.
Talk by J. Eisen for NZ Computational Genomics meeting

This document discusses phylogeny-driven approaches to studying microbial diversity using ribosomal RNA gene sequences. It provides background on how advances in sequencing technology and appreciation of microbial diversity have enabled microbiome research. The document outlines several uses of phylogeny in microbiome studies, including constructing species phylogenies using rRNA sequences and assigning taxonomy to environmental sequences via rRNA phylotyping. It describes challenges with analyzing large rRNA datasets and introduces an automated pipeline called STAP that generates high-quality multiple sequence alignments and phylogenetic trees to classify sequences and analyze species diversity in a manner that scales to large datasets.
Proteomics & Metabolomics

The Proteomics and Metabolomics Shared Resource (PMSR) at Georgetown University provides proteomics and metabolomics services and expertise. The document describes the PMSR's integrated proteomics workflow including 2D gel-based analysis, DIGE, spot picking, and MALDI TOF/TOF mass spectrometry for protein identification. Liquid chromatography-based proteomics using iTRAQ/ICAT labeling for quantitative analysis and a nano LC-QSTAR ELITE mass spectrometer are also described. The PMSR supports small molecule profiling and quantitation using UPLC-TOFMS and metabolomics applications.
Exploiting Edinburgh's Guide to PHARMACOLOGY database as a source of protein ...

Presented by Jamie Davies at the SULSA Synthetic Biology Meeting, Edinburgh, 10 June 2014
http://www.eventbrite.co.uk/e/sulsa-synthetic-biology-meeting-registration-11251454403?aff=eorg
Abstract: Synthetic creation of new biological systems typically incorporates pathways and signaling modules from known protein building blocks. Testing the models underpinning the synthetic engineering thus needs the experimental manipulation of individual proteins, for example, ablating a specific enzyme activity via RNAi, SNP mutation, or knockout. However, the option of small-molecule inhibition as the system perturbation has the advantages of 1) rapid onset 2) dose-response 3) analog testing for structure-activity relationships, 4) exploring mixtures for combinatorial effects 5) pulsing and reversal by wash-out. 6) accurate measurements of added substances and 7) a vast precedent of published results in natural systems from medicinal chemistry, pharmacology, and chemical biology. For the synthetic biologists the GToPdb1 can thus be considered as compendium of the latter. It encompasses an interaction matrix between ~4000 small molecules and ~1000 human proteins with a focus on drugs, clinical candidates, research compounds and peptide ligands These not only have ~ 10,000 mapped binding constants but also the spectrum of documented modulation extends across enzymes, receptors, channels and transporters. It thus becomes an increasingly plausible option to choose a “Lego protein” from GToPdb as a synthetic system component that can have experimentally useable activity probes available from chemical vendors. Even if it does not currently have a suitable target-probe pair, as knowledge base (and expertise resource via the curation team who populate it) GToPdb is an ideal starting point from which to walk out to wider chemogenomic spaces. For example, while an approved drug and its target might seem a logical choice, analogs from the lead series or different chemotypes from which the drug was optimized, or even failed in development, can have superior probe-like properties for in vitro experiments (e.g. be more potent, specific and soluble). The GToPdb facilitates access to such compound data via curated papers and patents.
References
1. Pawson AJ, Sharman JL, Benson HE, Faccenda E, Alexander SP, Buneman OP, Davenport AP, McGrath JC, Peters JA, Southan C, Spedding M, Yu W, Harmar AJ; NC-IUPHAR. The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 2014 Jan 1;42(1)
Identification of PFOA linked metabolic diseases by crossing databases

The increasing amount of biological data makes possible their interpretation more accurate and richer than never before. Various way of representations and interpretations of the links between those data have been applied or developed consequently to these new elements which can be taken into account in diagnostics and soon in personalized medicine. The aim of this student project was to cross data coming from various databases to be able to link Perfluorooctaoic Acid (PFOA) to one or more human phenotypes and metabolic diseases. Our approach makes possible an easy and confident interpretation on the data kept and also allow us to rank diseases linked according to their risk of correlation to a specific set of proteins.
Effect of Food Source on Enzymatic Activity in C. maculatus [draft 2]![Effect of Food Source on Enzymatic Activity in C. maculatus [draft 2]](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Effect of Food Source on Enzymatic Activity in C. maculatus [draft 2]](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
1. The study investigated the effect of food source (mung beans vs cowpeas) on enzymatic activity in Callosobruchus maculatus beetles, which are agricultural pests.
2. Experiments measured alpha-naphthyl acetate esterase (ANAE) and beta-naphthyl acetate esterase (BNAE) activity in beetles fed each food source.
3. Statistical analysis using t-tests found no significant differences in ANAE or BNAE activity between the food sources, failing to support the hypothesis that food source affects enzymatic activity.
Building an efficient infrastructure, standards and data flow for metabolomics

Christoph Steinbeck from the European Bioinformatics Institute discusses developing efficient infrastructure, standards, and data flow for metabolomics. Metabolomics measures small molecule metabolites in organisms and generates large amounts of data. The MetaboLights database was created to share metabolomics data openly. It is growing rapidly with a doubling time of 3 months for metabolomics data. Efforts are underway to build standardized reference data through projects like COSMOS and MetabolomeXchange. While genomes of thousands of species have been sequenced, far fewer complete metabolomes exist. The talk advocates for focused efforts to map metabolites and build quantitative models of well-studied model organisms' metabolomes.
ADARSH JOSE_Resume

Adarsh Jose is a PhD candidate in Bioinformatics and Computational Biology at Iowa State University. His research focuses on analyzing next-generation sequencing data to study gene expression, metabolic pathways, and protein interactions in various organisms. He has developed software for functional annotation and ortholog prediction from transcriptomes. Jose has presented his work at several conferences and collaborates with researchers studying hydrocarbon synthesis in microbes and plants.
Recommended
CECAM-2015_ESR10_poster_Andrew-Brockman

This research aims to identify protein-protein interactions in Anopheles gambiae that could be targeted by ligands to block transmission of malaria. Computational methods were used to predict nearly 10,000 putative interactions and 100 interaction modules. The interactions were identified using various data sources and techniques including gene expression, regulatory motifs, orthology, and literature mining. Validation with additional methods like structural analysis could help confirm some of the predicted interactions. The goal is to develop novel drugs, insecticides or repellents by disrupting interactions essential for malaria transmission.
Talk by J. Eisen for NZ Computational Genomics meeting

This document discusses phylogeny-driven approaches to studying microbial diversity using ribosomal RNA gene sequences. It provides background on how advances in sequencing technology and appreciation of microbial diversity have enabled microbiome research. The document outlines several uses of phylogeny in microbiome studies, including constructing species phylogenies using rRNA sequences and assigning taxonomy to environmental sequences via rRNA phylotyping. It describes challenges with analyzing large rRNA datasets and introduces an automated pipeline called STAP that generates high-quality multiple sequence alignments and phylogenetic trees to classify sequences and analyze species diversity in a manner that scales to large datasets.
Proteomics & Metabolomics

The Proteomics and Metabolomics Shared Resource (PMSR) at Georgetown University provides proteomics and metabolomics services and expertise. The document describes the PMSR's integrated proteomics workflow including 2D gel-based analysis, DIGE, spot picking, and MALDI TOF/TOF mass spectrometry for protein identification. Liquid chromatography-based proteomics using iTRAQ/ICAT labeling for quantitative analysis and a nano LC-QSTAR ELITE mass spectrometer are also described. The PMSR supports small molecule profiling and quantitation using UPLC-TOFMS and metabolomics applications.
Exploiting Edinburgh's Guide to PHARMACOLOGY database as a source of protein ...

Presented by Jamie Davies at the SULSA Synthetic Biology Meeting, Edinburgh, 10 June 2014
http://www.eventbrite.co.uk/e/sulsa-synthetic-biology-meeting-registration-11251454403?aff=eorg
Abstract: Synthetic creation of new biological systems typically incorporates pathways and signaling modules from known protein building blocks. Testing the models underpinning the synthetic engineering thus needs the experimental manipulation of individual proteins, for example, ablating a specific enzyme activity via RNAi, SNP mutation, or knockout. However, the option of small-molecule inhibition as the system perturbation has the advantages of 1) rapid onset 2) dose-response 3) analog testing for structure-activity relationships, 4) exploring mixtures for combinatorial effects 5) pulsing and reversal by wash-out. 6) accurate measurements of added substances and 7) a vast precedent of published results in natural systems from medicinal chemistry, pharmacology, and chemical biology. For the synthetic biologists the GToPdb1 can thus be considered as compendium of the latter. It encompasses an interaction matrix between ~4000 small molecules and ~1000 human proteins with a focus on drugs, clinical candidates, research compounds and peptide ligands These not only have ~ 10,000 mapped binding constants but also the spectrum of documented modulation extends across enzymes, receptors, channels and transporters. It thus becomes an increasingly plausible option to choose a “Lego protein” from GToPdb as a synthetic system component that can have experimentally useable activity probes available from chemical vendors. Even if it does not currently have a suitable target-probe pair, as knowledge base (and expertise resource via the curation team who populate it) GToPdb is an ideal starting point from which to walk out to wider chemogenomic spaces. For example, while an approved drug and its target might seem a logical choice, analogs from the lead series or different chemotypes from which the drug was optimized, or even failed in development, can have superior probe-like properties for in vitro experiments (e.g. be more potent, specific and soluble). The GToPdb facilitates access to such compound data via curated papers and patents.
References
1. Pawson AJ, Sharman JL, Benson HE, Faccenda E, Alexander SP, Buneman OP, Davenport AP, McGrath JC, Peters JA, Southan C, Spedding M, Yu W, Harmar AJ; NC-IUPHAR. The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 2014 Jan 1;42(1)
Identification of PFOA linked metabolic diseases by crossing databases

The increasing amount of biological data makes possible their interpretation more accurate and richer than never before. Various way of representations and interpretations of the links between those data have been applied or developed consequently to these new elements which can be taken into account in diagnostics and soon in personalized medicine. The aim of this student project was to cross data coming from various databases to be able to link Perfluorooctaoic Acid (PFOA) to one or more human phenotypes and metabolic diseases. Our approach makes possible an easy and confident interpretation on the data kept and also allow us to rank diseases linked according to their risk of correlation to a specific set of proteins.
Effect of Food Source on Enzymatic Activity in C. maculatus [draft 2]![Effect of Food Source on Enzymatic Activity in C. maculatus [draft 2]](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Effect of Food Source on Enzymatic Activity in C. maculatus [draft 2]](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
1. The study investigated the effect of food source (mung beans vs cowpeas) on enzymatic activity in Callosobruchus maculatus beetles, which are agricultural pests.
2. Experiments measured alpha-naphthyl acetate esterase (ANAE) and beta-naphthyl acetate esterase (BNAE) activity in beetles fed each food source.
3. Statistical analysis using t-tests found no significant differences in ANAE or BNAE activity between the food sources, failing to support the hypothesis that food source affects enzymatic activity.
Building an efficient infrastructure, standards and data flow for metabolomics

Christoph Steinbeck from the European Bioinformatics Institute discusses developing efficient infrastructure, standards, and data flow for metabolomics. Metabolomics measures small molecule metabolites in organisms and generates large amounts of data. The MetaboLights database was created to share metabolomics data openly. It is growing rapidly with a doubling time of 3 months for metabolomics data. Efforts are underway to build standardized reference data through projects like COSMOS and MetabolomeXchange. While genomes of thousands of species have been sequenced, far fewer complete metabolomes exist. The talk advocates for focused efforts to map metabolites and build quantitative models of well-studied model organisms' metabolomes.
ADARSH JOSE_Resume

Adarsh Jose is a PhD candidate in Bioinformatics and Computational Biology at Iowa State University. His research focuses on analyzing next-generation sequencing data to study gene expression, metabolic pathways, and protein interactions in various organisms. He has developed software for functional annotation and ortholog prediction from transcriptomes. Jose has presented his work at several conferences and collaborates with researchers studying hydrocarbon synthesis in microbes and plants.
Ransbotyn et al PUBLISHED (1)

This study developed a computational approach combining gene expression ranking and co-expression network analysis to identify novel stress regulatory genes in Arabidopsis thaliana. The researchers ranked genes based on their expression response to multiple abiotic stresses and analyzed co-expression networks to select candidate stress regulatory genes. Screening 62 mutants defective in candidate genes yielded a remarkably high gene discovery rate of up to 62% for stress phenotypes, far greater than typical screens. Additionally screening 64 other mutants based only on expression ranking yielded a lower but still improved rate of 36%, showing the power of combining methods. This systems approach can enhance gene discovery for other processes given suitable transcriptome data.
Gdt 2-126 (1)

1) The document discusses interfaces between nanoelectronic and biological systems, including applications of nanotechnology in medicine (nanomedicine) and molecular biomimetics.
2) It describes techniques like phage display and cell surface display that use libraries of random peptides to select sequences that bind to inorganic surfaces for constructing hybrid biomaterials.
3) The applications discussed include bio-nanowire devices for ultrasensitive detection, nanowire-based sensors for cancer markers, and interfaces between nanoelectronic devices and cells.
Gdt 2-126

1) Nanomedicine uses nanotechnology to prevent and treat disease by imparting properties like antibacterial and anti-odor functionality to skin products using nanomaterials.
2) Molecular biomimetics marries materials science and molecular biology to develop functional hybrid systems composed of inorganic and protein materials. It uses protein templates designed through genetics, surface proteins to bind synthetic entities, and the ability of proteins to self-assemble into complex structures.
3) Combinatorial biology techniques like phage display and cell-surface display are used to select protein sequences that strongly bind to inorganic surfaces from large random peptide libraries. This allows obtaining inorganic-binding proteins without an a priori knowledge of their sequence.
Semantic (Web) Technologies for Translational Research in Life Sciences

Semantic (Web) Technologies for Translational Research in Life SciencesArtificial Intelligence Institute at UofSC
This document discusses using semantic web technologies for translational research in life sciences. It provides an overview of semantic web standards and outlines several projects demonstrating applications in healthcare and biomedical research. These include developing an active semantic electronic medical record, semantically annotating experimental glycomics data, and integrating diverse biomedical data sources using ontologies to enable complex querying and knowledge discovery.Metagenomics and it’s applications

Shotgun metagenomics sequencing allows researchers to comprehensively sample all genes in organisms present in a complex sample without culturing. This provides insights into bacterial diversity, abundance, and uncultured microbes. Bioinformatics pipelines guide analysis including quality filtering, assembly, binning, gene finding, fingerprinting, and phylogeny/diversity modeling to understand communities. Metagenomics has applications in antibiotic/drug discovery, bioremediation, agriculture, human microbiome mapping, and more. Tools like QIIME, Mothur, MEGAN, and MG-RAST facilitate large-scale metagenomic analysis.
metagenomicsanditsapplications-161222180924.pdf

Shotgun metagenomics sequencing allows researchers to comprehensively sample all genes in organisms present in a complex sample without relying on cultivation. This approach provides insights into bacterial diversity, abundance, and unculturable microbes. Bioinformatics pipelines guide the optimized whole genome shotgun sequencing approach by performing tasks like quality control, assembly, binning, gene finding, fingerprinting, and phylogenetic analysis to study community diversity from fragmented metagenomic data. Metagenomics has applications in fields like drug discovery, bioremediation, agriculture, and understanding the human microbiome.
B.3.5

The document discusses using WEKA and BioWeka to analyze DNA sequences and perform pattern matching. It summarizes how Eclat filtering and EM clustering are applied to a dataset containing DNA sequences from human and chimpanzee chromosomes. Eclat is used to extract codon frequencies as features, while EM clustering assigns sequences to clusters based on the mixture model with the highest posterior probability. The analysis aims to identify biologically relevant groups of genes and determine chromosomal similarities between humans and chimpanzees.
Current advances to bridge the usability-expressivity gap in biomedical seman...

I presented a talk at the Protege research meeting on the 'Current advances to bridge the usability-expressivity gap in biomedical semantic search (and visualizing linked data)' https://sites.google.com/site/protegeresearchmeeting/meeting-materials/current-advances-to-bridge-the-usability-expressivity-gap-in-semantic-search
Resume-Cover letter-Ali Ashrafzadeh020416

The document provides a summary of Ali Ashrafzadeh's background and qualifications. He has over 13 years of experience in molecular research and teaching at veterinary schools. His expertise includes mass spectrometry, proteomics, metabolomics, semen quality evaluation, and experimental design. He has a PhD in Animal Biotechnology and has worked in various roles involving running and maintaining mass spectrometry instruments, collaborating on research projects, mentoring students, and publishing findings.
Maize database 

Maize database is the most important database in the bioinformatics. so i hope it is beneficial to the B.Sc.in Agriculture and M.Sc. in Genetics and Plant Breeding.
A Critical Assessment Of Mus Musculus Gene Function Prediction Using Integrat...

This study assessed methods for predicting gene function in mice using integrated genomic data. Researchers provided standardized mouse genomic and functional annotation data to 9 bioinformatics teams. The teams used this data to independently train classifiers and predict functions, defined by Gene Ontology terms, for over 21,000 mouse genes. The best performing predictions were combined. This approach inferred functions for 76% of mouse genes, including 5,000 previously uncharacterized genes. At a 20% recall rate, the unified predictions averaged 41% precision, with 26% of terms achieving over 90% precision. The results demonstrate that currently available mammalian data allows predicting gene functions with both breadth and accuracy, including many novel predictions for previously uncharacterized genes.
Roleoffunctionalgenomicsincropimprovement ashishgautam

Functional genomics is a general approach toward understanding how the genes of an organism work together by assigning new functions to unknown genes. Information about the hypothesized function of an unknown gene may be deduced from its sequence structure using already known functions of similar genes as the basis for comparison. Gene function analysis therefore necessitates the analysis of temporal and spatial gene expression patterns (Yunbi Xu et al , Plant Molecular Biology (2005) ).
FAIR Agronomy, where are we? The KnetMiner Use Case

Marco Brandizi and Keywan Hassani-Pak, Rothamsted Research, Invited Presentation at SWAT4HCLS 2022.
FAIR data principles are being a driving force in life sciences and other scientific domains, helping researchers to share their data and free all of their potential to integrate information and do novel discoveries. Knowledge graphs are an ever more popular paradigm to model data according to such principles, and technologies such as graph databases are emerging as complementary to approaches like linked data. All of this includes the agronomy, farming and food domains. How advanced the adoption of sound data management policies is in these life domains? How does that compare to other life sciences? In this presentation, we will talk about our practical experience, focusing on KnetMiner, a gene and molecular biology discovering platform, which is based on building and publishing knowledge graphs according to the FAIR principles, as well as using a mix of linked data standards for life sciences and recent graph database and API technologies. We will welcome questions and discussions from the audience about similar experience.
2015 mcgill-talk

The document discusses strategies for working with large biological datasets as sequencing costs decrease and data volumes increase exponentially. It summarizes three key uses for abundant sequencing data: hypothesis falsification, model comparison, and hypothesis generation. The author's lab aims to develop open tools for moving quickly from raw data to hypotheses and identify challenges preventing collaborators from doing their science. Summarizing a discussion on soil microbial communities, it notes the immense diversity and challenges of culture-dependent approaches, necessitating single-cell sequencing and metagenomics.
Thesis def

Jays thesis defense on database federation, data marts and the power of integrated protein bioinformatics.
Chapter 20 ppt

This document summarizes key concepts from Chapter 20 of an AP Biology textbook. It discusses several topics:
1) Genomics is the study of genomes and how they are organized and regulated. Genome sequences provide insights into fundamental biological questions.
2) Computer analysis can identify protein-coding genes in DNA sequences by looking for start/stop signals and other features. With 25,000 genes in humans, this analysis is a huge undertaking without technology.
3) Genome sizes vary greatly between organisms, but size does not always correlate with complexity. Some plants have genomes much larger than humans despite fewer genes.
MORPH-R article

This document describes MORPH-R, a tool for predicting missing genes in biological pathways. MORPH-R is available both as a web server and standalone software. It uses the MORPH algorithm to rank candidate genes for their likelihood of participating in or affecting biological pathways of interest. MORPH-R was tested on potato data and achieved high performance, identifying novel candidate genes for potato pathways and gene ontology categories. The tool allows users to analyze pathways in tomato, Arabidopsis, rice and potato, and can be customized to analyze additional organisms by adding new gene expression and interaction network data.
rheumatoid arthritis

This document provides an overview of bioinformatics and genomics. It begins with an acknowledgement and abstract section. The introduction defines bioinformatics and its role in analyzing genetic sequences and biological data through computational methods. Major research areas of bioinformatics discussed include sequence analysis, genome annotation, evolutionary biology, measuring biodiversity, gene expression analysis, protein analysis, cancer mutation analysis, and protein structure prediction. Comparative genomics and modeling biological systems are also summarized. The document concludes with a definition of genomics as the study of genomes through sequencing efforts and mapping genetic interactions.
Munoz torres web-apollo-workshop_exeter-2014_ss

This document provides an introduction and overview of a manual annotation workshop using the Web Apollo genome annotation tool. It discusses manual annotation and community-based curation efforts. The workshop aims to teach participants how to identify genes of interest, become familiar with Web Apollo, learn how to corroborate and modify gene models using evidence, and understand the genome annotation process from assembly to manual curation. The document outlines the workshop activities and provides guidance on using Web Apollo, including navigating the interface, editing annotations, and annotating simple cases by adding or modifying exons.
Biological Significance of Gene Expression Data Using Similarity Based Biclus...

Unlocking the complexity of a living organism’s biological processes, functions and genetic network is vital in learning how to improve the health of humankind. Genetic analysis, especially biclustering, is a significant step in this process. Though many biclustering methods exist, only few provide a query based approach for biologists to search the biclusters which contain a certain gene of interest. This proposed query based biclustering algorithm SIMBIC+ first identifies a functionally rich query gene. After identifying the query gene, sets of genes including query gene that show coherent expression patterns across subsets of experimental conditions is identified. It performs simultaneous clustering on both row and column dimension to extract biclusters using Top down approach. Since it uses novel ‘ratio’ based similarity measure, biclusters with more coherence and with more biological meaning are identified. SIMBIC+ uses score based approach with an aim of maximizing the similarity of the bicluster. Contribution entropy based condition selection and multiple row / column deletion methods are used to reduce the complexity of the algorithm to identify biclusters with maximum similarity value. Experiments are conducted on Yeast Saccharomyces dataset and the biclusters obtained are compared with biclusters of popular MSB (Maximum Similarity Bicluster) algorithm. The biological significance of the biclusters obtained by the proposed algorithm and MSB are compared and the comparison proves that SIMBIC+ identifies biclusters with more significant GO (Gene Ontology).
seed production, Nursery & Gardening.pdf

This presentation offers a general idea of the structure of seed, seed production, management of seeds and its allied technologies. It also offers the concept of gene erosion and the practices used to control it. Nursery and gardening have been widely explored along with their importance in the related domain.
Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf![Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Music and Medieval History
More Related Content
Similar to Integrated approaches for the discovery of novel enzymatic activities
Ransbotyn et al PUBLISHED (1)

This study developed a computational approach combining gene expression ranking and co-expression network analysis to identify novel stress regulatory genes in Arabidopsis thaliana. The researchers ranked genes based on their expression response to multiple abiotic stresses and analyzed co-expression networks to select candidate stress regulatory genes. Screening 62 mutants defective in candidate genes yielded a remarkably high gene discovery rate of up to 62% for stress phenotypes, far greater than typical screens. Additionally screening 64 other mutants based only on expression ranking yielded a lower but still improved rate of 36%, showing the power of combining methods. This systems approach can enhance gene discovery for other processes given suitable transcriptome data.
Gdt 2-126 (1)

1) The document discusses interfaces between nanoelectronic and biological systems, including applications of nanotechnology in medicine (nanomedicine) and molecular biomimetics.
2) It describes techniques like phage display and cell surface display that use libraries of random peptides to select sequences that bind to inorganic surfaces for constructing hybrid biomaterials.
3) The applications discussed include bio-nanowire devices for ultrasensitive detection, nanowire-based sensors for cancer markers, and interfaces between nanoelectronic devices and cells.
Gdt 2-126

1) Nanomedicine uses nanotechnology to prevent and treat disease by imparting properties like antibacterial and anti-odor functionality to skin products using nanomaterials.
2) Molecular biomimetics marries materials science and molecular biology to develop functional hybrid systems composed of inorganic and protein materials. It uses protein templates designed through genetics, surface proteins to bind synthetic entities, and the ability of proteins to self-assemble into complex structures.
3) Combinatorial biology techniques like phage display and cell-surface display are used to select protein sequences that strongly bind to inorganic surfaces from large random peptide libraries. This allows obtaining inorganic-binding proteins without an a priori knowledge of their sequence.
Semantic (Web) Technologies for Translational Research in Life Sciences

Semantic (Web) Technologies for Translational Research in Life SciencesArtificial Intelligence Institute at UofSC
This document discusses using semantic web technologies for translational research in life sciences. It provides an overview of semantic web standards and outlines several projects demonstrating applications in healthcare and biomedical research. These include developing an active semantic electronic medical record, semantically annotating experimental glycomics data, and integrating diverse biomedical data sources using ontologies to enable complex querying and knowledge discovery.Metagenomics and it’s applications

Shotgun metagenomics sequencing allows researchers to comprehensively sample all genes in organisms present in a complex sample without culturing. This provides insights into bacterial diversity, abundance, and uncultured microbes. Bioinformatics pipelines guide analysis including quality filtering, assembly, binning, gene finding, fingerprinting, and phylogeny/diversity modeling to understand communities. Metagenomics has applications in antibiotic/drug discovery, bioremediation, agriculture, human microbiome mapping, and more. Tools like QIIME, Mothur, MEGAN, and MG-RAST facilitate large-scale metagenomic analysis.
metagenomicsanditsapplications-161222180924.pdf

Shotgun metagenomics sequencing allows researchers to comprehensively sample all genes in organisms present in a complex sample without relying on cultivation. This approach provides insights into bacterial diversity, abundance, and unculturable microbes. Bioinformatics pipelines guide the optimized whole genome shotgun sequencing approach by performing tasks like quality control, assembly, binning, gene finding, fingerprinting, and phylogenetic analysis to study community diversity from fragmented metagenomic data. Metagenomics has applications in fields like drug discovery, bioremediation, agriculture, and understanding the human microbiome.
B.3.5

The document discusses using WEKA and BioWeka to analyze DNA sequences and perform pattern matching. It summarizes how Eclat filtering and EM clustering are applied to a dataset containing DNA sequences from human and chimpanzee chromosomes. Eclat is used to extract codon frequencies as features, while EM clustering assigns sequences to clusters based on the mixture model with the highest posterior probability. The analysis aims to identify biologically relevant groups of genes and determine chromosomal similarities between humans and chimpanzees.
Current advances to bridge the usability-expressivity gap in biomedical seman...

I presented a talk at the Protege research meeting on the 'Current advances to bridge the usability-expressivity gap in biomedical semantic search (and visualizing linked data)' https://sites.google.com/site/protegeresearchmeeting/meeting-materials/current-advances-to-bridge-the-usability-expressivity-gap-in-semantic-search
Resume-Cover letter-Ali Ashrafzadeh020416

The document provides a summary of Ali Ashrafzadeh's background and qualifications. He has over 13 years of experience in molecular research and teaching at veterinary schools. His expertise includes mass spectrometry, proteomics, metabolomics, semen quality evaluation, and experimental design. He has a PhD in Animal Biotechnology and has worked in various roles involving running and maintaining mass spectrometry instruments, collaborating on research projects, mentoring students, and publishing findings.
Maize database 

Maize database is the most important database in the bioinformatics. so i hope it is beneficial to the B.Sc.in Agriculture and M.Sc. in Genetics and Plant Breeding.
A Critical Assessment Of Mus Musculus Gene Function Prediction Using Integrat...

This study assessed methods for predicting gene function in mice using integrated genomic data. Researchers provided standardized mouse genomic and functional annotation data to 9 bioinformatics teams. The teams used this data to independently train classifiers and predict functions, defined by Gene Ontology terms, for over 21,000 mouse genes. The best performing predictions were combined. This approach inferred functions for 76% of mouse genes, including 5,000 previously uncharacterized genes. At a 20% recall rate, the unified predictions averaged 41% precision, with 26% of terms achieving over 90% precision. The results demonstrate that currently available mammalian data allows predicting gene functions with both breadth and accuracy, including many novel predictions for previously uncharacterized genes.
Roleoffunctionalgenomicsincropimprovement ashishgautam

Functional genomics is a general approach toward understanding how the genes of an organism work together by assigning new functions to unknown genes. Information about the hypothesized function of an unknown gene may be deduced from its sequence structure using already known functions of similar genes as the basis for comparison. Gene function analysis therefore necessitates the analysis of temporal and spatial gene expression patterns (Yunbi Xu et al , Plant Molecular Biology (2005) ).
FAIR Agronomy, where are we? The KnetMiner Use Case

Marco Brandizi and Keywan Hassani-Pak, Rothamsted Research, Invited Presentation at SWAT4HCLS 2022.
FAIR data principles are being a driving force in life sciences and other scientific domains, helping researchers to share their data and free all of their potential to integrate information and do novel discoveries. Knowledge graphs are an ever more popular paradigm to model data according to such principles, and technologies such as graph databases are emerging as complementary to approaches like linked data. All of this includes the agronomy, farming and food domains. How advanced the adoption of sound data management policies is in these life domains? How does that compare to other life sciences? In this presentation, we will talk about our practical experience, focusing on KnetMiner, a gene and molecular biology discovering platform, which is based on building and publishing knowledge graphs according to the FAIR principles, as well as using a mix of linked data standards for life sciences and recent graph database and API technologies. We will welcome questions and discussions from the audience about similar experience.
2015 mcgill-talk

The document discusses strategies for working with large biological datasets as sequencing costs decrease and data volumes increase exponentially. It summarizes three key uses for abundant sequencing data: hypothesis falsification, model comparison, and hypothesis generation. The author's lab aims to develop open tools for moving quickly from raw data to hypotheses and identify challenges preventing collaborators from doing their science. Summarizing a discussion on soil microbial communities, it notes the immense diversity and challenges of culture-dependent approaches, necessitating single-cell sequencing and metagenomics.
Thesis def

Jays thesis defense on database federation, data marts and the power of integrated protein bioinformatics.
Chapter 20 ppt

This document summarizes key concepts from Chapter 20 of an AP Biology textbook. It discusses several topics:
1) Genomics is the study of genomes and how they are organized and regulated. Genome sequences provide insights into fundamental biological questions.
2) Computer analysis can identify protein-coding genes in DNA sequences by looking for start/stop signals and other features. With 25,000 genes in humans, this analysis is a huge undertaking without technology.
3) Genome sizes vary greatly between organisms, but size does not always correlate with complexity. Some plants have genomes much larger than humans despite fewer genes.
MORPH-R article

This document describes MORPH-R, a tool for predicting missing genes in biological pathways. MORPH-R is available both as a web server and standalone software. It uses the MORPH algorithm to rank candidate genes for their likelihood of participating in or affecting biological pathways of interest. MORPH-R was tested on potato data and achieved high performance, identifying novel candidate genes for potato pathways and gene ontology categories. The tool allows users to analyze pathways in tomato, Arabidopsis, rice and potato, and can be customized to analyze additional organisms by adding new gene expression and interaction network data.
rheumatoid arthritis

This document provides an overview of bioinformatics and genomics. It begins with an acknowledgement and abstract section. The introduction defines bioinformatics and its role in analyzing genetic sequences and biological data through computational methods. Major research areas of bioinformatics discussed include sequence analysis, genome annotation, evolutionary biology, measuring biodiversity, gene expression analysis, protein analysis, cancer mutation analysis, and protein structure prediction. Comparative genomics and modeling biological systems are also summarized. The document concludes with a definition of genomics as the study of genomes through sequencing efforts and mapping genetic interactions.
Munoz torres web-apollo-workshop_exeter-2014_ss

This document provides an introduction and overview of a manual annotation workshop using the Web Apollo genome annotation tool. It discusses manual annotation and community-based curation efforts. The workshop aims to teach participants how to identify genes of interest, become familiar with Web Apollo, learn how to corroborate and modify gene models using evidence, and understand the genome annotation process from assembly to manual curation. The document outlines the workshop activities and provides guidance on using Web Apollo, including navigating the interface, editing annotations, and annotating simple cases by adding or modifying exons.
Biological Significance of Gene Expression Data Using Similarity Based Biclus...

Unlocking the complexity of a living organism’s biological processes, functions and genetic network is vital in learning how to improve the health of humankind. Genetic analysis, especially biclustering, is a significant step in this process. Though many biclustering methods exist, only few provide a query based approach for biologists to search the biclusters which contain a certain gene of interest. This proposed query based biclustering algorithm SIMBIC+ first identifies a functionally rich query gene. After identifying the query gene, sets of genes including query gene that show coherent expression patterns across subsets of experimental conditions is identified. It performs simultaneous clustering on both row and column dimension to extract biclusters using Top down approach. Since it uses novel ‘ratio’ based similarity measure, biclusters with more coherence and with more biological meaning are identified. SIMBIC+ uses score based approach with an aim of maximizing the similarity of the bicluster. Contribution entropy based condition selection and multiple row / column deletion methods are used to reduce the complexity of the algorithm to identify biclusters with maximum similarity value. Experiments are conducted on Yeast Saccharomyces dataset and the biclusters obtained are compared with biclusters of popular MSB (Maximum Similarity Bicluster) algorithm. The biological significance of the biclusters obtained by the proposed algorithm and MSB are compared and the comparison proves that SIMBIC+ identifies biclusters with more significant GO (Gene Ontology).
Similar to Integrated approaches for the discovery of novel enzymatic activities (20)
Semantic (Web) Technologies for Translational Research in Life Sciences

Semantic (Web) Technologies for Translational Research in Life Sciences
Current advances to bridge the usability-expressivity gap in biomedical seman...

Current advances to bridge the usability-expressivity gap in biomedical seman...
A Critical Assessment Of Mus Musculus Gene Function Prediction Using Integrat...

A Critical Assessment Of Mus Musculus Gene Function Prediction Using Integrat...
Roleoffunctionalgenomicsincropimprovement ashishgautam

Roleoffunctionalgenomicsincropimprovement ashishgautam
FAIR Agronomy, where are we? The KnetMiner Use Case

FAIR Agronomy, where are we? The KnetMiner Use Case
Biological Significance of Gene Expression Data Using Similarity Based Biclus...

Biological Significance of Gene Expression Data Using Similarity Based Biclus...
Recently uploaded
seed production, Nursery & Gardening.pdf

This presentation offers a general idea of the structure of seed, seed production, management of seeds and its allied technologies. It also offers the concept of gene erosion and the practices used to control it. Nursery and gardening have been widely explored along with their importance in the related domain.
Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf![Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Music and Medieval History
SAP Unveils Generative AI Innovations at Annual Sapphire Conference

At its annual SAP Sapphire conference, SAP introduced groundbreaking generative AI advancements and strategic partnerships, underscoring its commitment to revolutionizing business operations in the AI era. By integrating Business AI throughout its enterprise cloud portfolio, which supports the world's most critical processes, SAP is fostering a new wave of business insight and creativity.
Embracing Deep Variability For Reproducibility and Replicability

Embracing Deep Variability For Reproducibility and ReplicabilityUniversity of Rennes, INSA Rennes, Inria/IRISA, CNRS
Embracing Deep Variability For Reproducibility and Replicability
Abstract: Reproducibility (aka determinism in some cases) constitutes a fundamental aspect in various fields of computer science, such as floating-point computations in numerical analysis and simulation, concurrency models in parallelism, reproducible builds for third parties integration and packaging, and containerization for execution environments. These concepts, while pervasive across diverse concerns, often exhibit intricate inter-dependencies, making it challenging to achieve a comprehensive understanding. In this short and vision paper we delve into the application of software engineering techniques, specifically variability management, to systematically identify and explicit points of variability that may give rise to reproducibility issues (eg language, libraries, compiler, virtual machine, OS, environment variables, etc). The primary objectives are: i) gaining insights into the variability layers and their possible interactions, ii) capturing and documenting configurations for the sake of reproducibility, and iii) exploring diverse configurations to replicate, and hence validate and ensure the robustness of results. By adopting these methodologies, we aim to address the complexities associated with reproducibility and replicability in modern software systems and environments, facilitating a more comprehensive and nuanced perspective on these critical aspects.
https://hal.science/hal-04582287
Post translation modification by Suyash Garg

overview of PTM helps to the students who wants to clear their basics about it.
JAMES WEBB STUDY THE MASSIVE BLACK HOLE SEEDS

The pathway(s) to seeding the massive black holes (MBHs) that exist at the heart of galaxies in the present and distant Universe remains an unsolved problem. Here we categorise, describe and quantitatively discuss the formation pathways of both light and heavy seeds. We emphasise that the most recent computational models suggest that rather than a bimodal-like mass spectrum between light and heavy seeds with light at one end and heavy at the other that instead a continuum exists. Light seeds being more ubiquitous and the heavier seeds becoming less and less abundant due the rarer environmental conditions required for their formation. We therefore examine the different mechanisms that give rise to different seed mass spectrums. We show how and why the mechanisms that produce the heaviest seeds are also among the rarest events in the Universe and are hence extremely unlikely to be the seeds for the vast majority of the MBH population. We quantify, within the limits of the current large uncertainties in the seeding processes, the expected number densities of the seed mass spectrum. We argue that light seeds must be at least 103 to 105 times more numerous than heavy seeds to explain the MBH population as a whole. Based on our current understanding of the seed population this makes heavy seeds (Mseed > 103 M⊙) a significantly more likely pathway given that heavy seeds have an abundance pattern than is close to and likely in excess of 10−4 compared to light seeds. Finally, we examine the current state-of-the-art in numerical calculations and recent observations and plot a path forward for near-future advances in both domains.
Rodents, Birds and locust_Pests of crops.pdf

Mole rat or Lesser bandicoot rat, Bandicotabengalensis
•Head -round and broad muzzle
•Tail -shorter than head, body
•Prefers damp areas
•Burrows with scooped soil before entrance
•Potential rat, one pair can produce more than 800 offspringsin one year
حبوب الاجهاض الامارات | 00971547952044 | حبوب اجهاض امارات للبيع

حبوب الاجهاض الامارات | 00971547952044 | حبوب اجهاض امارات للبيعحبوب الاجهاض الامارات حبوب سايتوتك الامارات
إتصل على هذا الرقم اذا اردت الحصول على "حبوب الاجهاض الامارات" توصيلنا مجاني رقم الواتساب 00971547952044:
00971547952044. حبوب الإجهاض في دبي | أبوظبي | الشارقة | السطوة | سعر سايتوتك Cytotec يتميز دواء Cytotec (سايتوتك) بفعاليته في إجهاض الحمل. يمكن الحصول على حبوب الاجهاض الامارات بسهولة من خلال خدمات التوصيل السريع والدفع عند الاستلام. تُستخدم حبوب سايتوتك بشكل شائع لإنهاء الحمل غير المرغوب فيه. حبوب الاجهاض الامارات هي الخيار الأمثل لمن يبحث عن طريقة آمنة وفعالة للإجهاض المنزلي.
تتوفر حبوب الاجهاض الامارات بأسعار تنافسية، ويمكنك الحصول على خصم كبير عند الشراء الآن. حبوب الاجهاض الامارات معروفة بقدرتها الفعالة على إنهاء الحمل في الشهر الأول أو الثاني. إذا كنت تبحث عن حبوب لتنزيل الحمل في الشهر الثاني أو الأول، فإن حبوب الاجهاض الامارات هي الخيار المثالي.
دواء سايتوتك يحتوي على المادة الفعالة ميزوبروستول، التي تُستخدم لإجهاض الحمل والتخلص من النزيف ما بعد الولادة. يمكنك الآن الحصول على حبوب سايتوتك للبيع في دبي وأبوظبي والشارقة من خلال الاتصال برقم 00971547952044. نسعى لتقديم أفضل الخدمات في مجال حبوب الاجهاض الامارات، مع توفير حبوب سايتوتك الأصلية بأفضل الأسعار.
إذا كنت في دبي، أبوظبي، الشارقة أو العين، يمكنك الحصول على حبوب الاجهاض الامارات بسهولة وأمان. نحن نضمن لك وصول الحبوب الأصلية بسرية تامة مع خيار الدفع عند الاستلام. حبوب الاجهاض الامارات هي الحل الفعال لإنهاء الحمل غير المرغوب فيه بطريقة آمنة.
تبحث العديد من النساء في الإمارات العربية المتحدة عن حبوب الاجهاض الامارات كبديل للعمليات الجراحية التي تتطلب وقتاً طويلاً وتكلفة عالية. بفضل حبوب الاجهاض الامارات، يمكنك الآن إنهاء الحمل بسلام وأمان في منزلك. نحن نوفر حبوب الاجهاض الامارات الأصلية من إنتاج شركة فايزر، مما يضمن لك الحصول على منتج فعال وآمن.
إذا كنت تبحث عن حبوب الاجهاض الامارات في العين، دبي، أو أبوظبي، يمكنك التواصل معنا عبر الواتس آب أو الاتصال على رقم 00971547952044 للحصول على التفاصيل حول كيفية الشراء والتوصيل. حبوب الاجهاض الامارات متوفرة بأسعار تنافسية، مع تقديم خصومات كبيرة عند الشراء بالجملة.
حبوب الاجهاض الامارات هي الخيار الأمثل لمن تبحث عن وسيلة آمنة وسريعة لإنهاء الحمل غير المرغوب فيه. تواصل معنا اليوم للحصول على حبوب الاجهاض الامارات الأصلية وتجنب أي مشاكل أو مضاعفات صحية.
في النهاية، لا تقلق بشأن الحبوب المقلدة أو الخطرة، فنحن نوفر لك حبوب الاجهاض الامارات الأصلية بأفضل الأسعار وخدمة التوصيل السريع والآمن. اتصل بنا الآن على 00971547952044 لتأكيد طلبك والحصول على حبوب الاجهاض الامارات التي تحتاجها. نحن هنا لمساعدتك وتقديم الدعم اللازم لضمان حصولك على الحل المناسب لمشكلتك.Call Girls Noida🔥9873777170🔥Gorgeous Escorts in Noida Available 24/7

Call Girls Noida🔥9873777170🔥Gorgeous Escorts in Noida Available 24/7
Discovery of Merging Twin Quasars at z=6.05

We report the discovery of two quasars at a redshift of z = 6.05 in the process of merging. They were
serendipitously discovered from the deep multiband imaging data collected by the Hyper Suprime-Cam (HSC)
Subaru Strategic Program survey. The quasars, HSC J121503.42−014858.7 (C1) and HSC J121503.55−014859.3
(C2), both have luminous (>1043 erg s−1
) Lyα emission with a clear broad component (full width at half
maximum >1000 km s−1
). The rest-frame ultraviolet (UV) absolute magnitudes are M1450 = − 23.106 ± 0.017
(C1) and −22.662 ± 0.024 (C2). Our crude estimates of the black hole masses provide log 8.1 0. ( ) M M BH = 3
in both sources. The two quasars are separated by 12 kpc in projected proper distance, bridged by a structure in the
rest-UV light suggesting that they are undergoing a merger. This pair is one of the most distant merging quasars
reported to date, providing crucial insight into galaxy and black hole build-up in the hierarchical structure
formation scenario. A companion paper will present the gas and dust properties captured by Atacama Large
Millimeter/submillimeter Array observations, which provide additional evidence for and detailed measurements of
the merger, and also demonstrate that the two sources are not gravitationally lensed images of a single quasar.
Unified Astronomy Thesaurus concepts: Double quasars (406); Quasars (1319); Reionization (1383); High-redshift
galaxies (734); Active galactic nuclei (16); Galaxy mergers (608); Supermassive black holes (1663)
Candidate young stellar objects in the S-cluster: Kinematic analysis of a sub...

Context. The observation of several L-band emission sources in the S cluster has led to a rich discussion of their nature. However, a definitive answer to the classification of the dusty objects requires an explanation for the detection of compact Doppler-shifted Brγ emission. The ionized hydrogen in combination with the observation of mid-infrared L-band continuum emission suggests that most of these sources are embedded in a dusty envelope. These embedded sources are part of the S-cluster, and their relationship to the S-stars is still under debate. To date, the question of the origin of these two populations has been vague, although all explanations favor migration processes for the individual cluster members. Aims. This work revisits the S-cluster and its dusty members orbiting the supermassive black hole SgrA* on bound Keplerian orbits from a kinematic perspective. The aim is to explore the Keplerian parameters for patterns that might imply a nonrandom distribution of the sample. Additionally, various analytical aspects are considered to address the nature of the dusty sources. Methods. Based on the photometric analysis, we estimated the individual H−K and K−L colors for the source sample and compared the results to known cluster members. The classification revealed a noticeable contrast between the S-stars and the dusty sources. To fit the flux-density distribution, we utilized the radiative transfer code HYPERION and implemented a young stellar object Class I model. We obtained the position angle from the Keplerian fit results; additionally, we analyzed the distribution of the inclinations and the longitudes of the ascending node. Results. The colors of the dusty sources suggest a stellar nature consistent with the spectral energy distribution in the near and midinfrared domains. Furthermore, the evaporation timescales of dusty and gaseous clumps in the vicinity of SgrA* are much shorter ( 2yr) than the epochs covered by the observations (≈15yr). In addition to the strong evidence for the stellar classification of the D-sources, we also find a clear disk-like pattern following the arrangements of S-stars proposed in the literature. Furthermore, we find a global intrinsic inclination for all dusty sources of 60 ± 20◦, implying a common formation process. Conclusions. The pattern of the dusty sources manifested in the distribution of the position angles, inclinations, and longitudes of the ascending node strongly suggests two different scenarios: the main-sequence stars and the dusty stellar S-cluster sources share a common formation history or migrated with a similar formation channel in the vicinity of SgrA*. Alternatively, the gravitational influence of SgrA* in combination with a massive perturber, such as a putative intermediate mass black hole in the IRS 13 cluster, forces the dusty objects and S-stars to follow a particular orbital arrangement. Key words. stars: black holes– stars: formation– Galaxy: center– galaxies: star formation
Mapping the Growth of Supermassive Black Holes as a Function of Galaxy Stella...

The growth of supermassive black holes is strongly linked to their galaxies. It has been shown that the population
mean black hole accretion rate (BHAR) primarily correlates with the galaxy stellar mass (Må) and redshift for the
general galaxy population. This work aims to provide the best measurements of BHAR as a function of Må and
redshift over ranges of 109.5 < Må < 1012 Me and z < 4. We compile an unprecedentedly large sample with 8000
active galactic nuclei (AGNs) and 1.3 million normal galaxies from nine high-quality survey fields following a
wedding cake design. We further develop a semiparametric Bayesian method that can reasonably estimate BHAR
and the corresponding uncertainties, even for sparsely populated regions in the parameter space. BHAR is
constrained by X-ray surveys sampling the AGN accretion power and UV-to-infrared multiwavelength surveys
sampling the galaxy population. Our results can independently predict the X-ray luminosity function (XLF) from
the galaxy stellar mass function (SMF), and the prediction is consistent with the observed XLF. We also try adding
external constraints from the observed SMF and XLF. We further measure BHAR for star-forming and quiescent
galaxies and show that star-forming BHAR is generally larger than or at least comparable to the quiescent BHAR.
Unified Astronomy Thesaurus concepts: Supermassive black holes (1663); X-ray active galactic nuclei (2035);
Galaxies (573)
SDSS1335+0728: The awakening of a ∼ 106M⊙ black hole⋆

Context. The early-type galaxy SDSS J133519.91+072807.4 (hereafter SDSS1335+0728), which had exhibited no prior optical variations during the preceding two decades, began showing significant nuclear variability in the Zwicky Transient Facility (ZTF) alert stream from December 2019 (as ZTF19acnskyy). This variability behaviour, coupled with the host-galaxy properties, suggests that SDSS1335+0728 hosts a ∼ 106M⊙ black hole (BH) that is currently in the process of ‘turning on’. Aims. We present a multi-wavelength photometric analysis and spectroscopic follow-up performed with the aim of better understanding the origin of the nuclear variations detected in SDSS1335+0728. Methods. We used archival photometry (from WISE, 2MASS, SDSS, GALEX, eROSITA) and spectroscopic data (from SDSS and LAMOST) to study the state of SDSS1335+0728 prior to December 2019, and new observations from Swift, SOAR/Goodman, VLT/X-shooter, and Keck/LRIS taken after its turn-on to characterise its current state. We analysed the variability of SDSS1335+0728 in the X-ray/UV/optical/mid-infrared range, modelled its spectral energy distribution prior to and after December 2019, and studied the evolution of its UV/optical spectra. Results. From our multi-wavelength photometric analysis, we find that: (a) since 2021, the UV flux (from Swift/UVOT observations) is four times brighter than the flux reported by GALEX in 2004; (b) since June 2022, the mid-infrared flux has risen more than two times, and the W1−W2 WISE colour has become redder; and (c) since February 2024, the source has begun showing X-ray emission. From our spectroscopic follow-up, we see that (i) the narrow emission line ratios are now consistent with a more energetic ionising continuum; (ii) broad emission lines are not detected; and (iii) the [OIII] line increased its flux ∼ 3.6 years after the first ZTF alert, which implies a relatively compact narrow-line-emitting region. Conclusions. We conclude that the variations observed in SDSS1335+0728 could be either explained by a ∼ 106M⊙ AGN that is just turning on or by an exotic tidal disruption event (TDE). If the former is true, SDSS1335+0728 is one of the strongest cases of an AGNobserved in the process of activating. If the latter were found to be the case, it would correspond to the longest and faintest TDE ever observed (or another class of still unknown nuclear transient). Future observations of SDSS1335+0728 are crucial to further understand its behaviour. Key words. galaxies: active– accretion, accretion discs– galaxies: individual: SDSS J133519.91+072807.4
Recently uploaded (20)
Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf![Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Holsinger, Bruce W. - Music, body and desire in medieval culture [2001].pdf
SAP Unveils Generative AI Innovations at Annual Sapphire Conference

SAP Unveils Generative AI Innovations at Annual Sapphire Conference
Embracing Deep Variability For Reproducibility and Replicability

Embracing Deep Variability For Reproducibility and Replicability
حبوب الاجهاض الامارات | 00971547952044 | حبوب اجهاض امارات للبيع

حبوب الاجهاض الامارات | 00971547952044 | حبوب اجهاض امارات للبيع
Call Girls Noida🔥9873777170🔥Gorgeous Escorts in Noida Available 24/7

Call Girls Noida🔥9873777170🔥Gorgeous Escorts in Noida Available 24/7
Candidate young stellar objects in the S-cluster: Kinematic analysis of a sub...

Candidate young stellar objects in the S-cluster: Kinematic analysis of a sub...
Mapping the Growth of Supermassive Black Holes as a Function of Galaxy Stella...

Mapping the Growth of Supermassive Black Holes as a Function of Galaxy Stella...
SDSS1335+0728: The awakening of a ∼ 106M⊙ black hole⋆

SDSS1335+0728: The awakening of a ∼ 106M⊙ black hole⋆
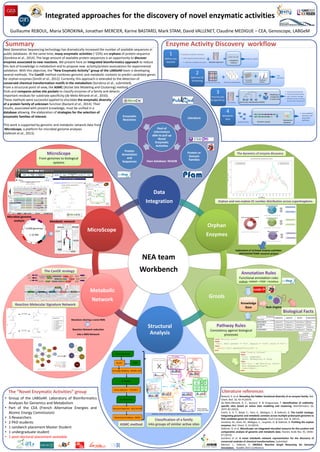
Integrated approaches for the discovery of novel enzymatic activities
- 1. Orphan and non-orphan EC number distribution across superkingdoms Integrated approaches for the discovery of novel enzymatic activities Guillaume REBOUL, Maria SOROKINA, Jonathan MERCIER, Karine BASTARD, Mark STAM, David VALLENET, Claudine MEDIGUE – CEA, Genoscope, LABGeM Annotation Rules Functional annotation rules UniRule: HAMAP + PIRSF + RuleBase Pathway Rules Consistency against biological processes Knowledge Base Rule Engine Enzyme Activity Discovery workflow Biological Facts rule "Missing state" when $fact: Fact( present == "no", require == "yes", avoid == "no" ) then modify( $fact.setState("missing") ); end rule "require Pathway" when $org: Organism() $path: Pathway(org == $org) then Fact fact = new Fact($org, $path); fact.setRequire("yes"); insert(fact); end The “Novel Enzymatic Activities” group • Group of the LABGeM: Laboratory of Bioinformatics Analyses for Genomics and Metabolism • Part of the CEA (French Alternative Energies and Atomic Energy Commission) • 3 Researchers • 2 PhD students • 1 sandwich placement Master Student • 1 undergraduate student • 1 post-doctoral placement available Pool of information able to pull up Novel Enzymatic Activities Protein or Domain Families Protein Annotation and Sequences Literature Enzymatic Reactions Own database: NEADB Summary ? iso-functional groups multi-functional group non-”activity” groups Motifs and key residues for function assignment 3 Promiscuity & Specificity Modeling of compounds in active sites + Full family new metabolic functions and associated pathways 4 Metabolic Role Biochemical validation + Genomic contextRepresentants enzymes family 1 Define one reaction +Multiple alignment one generic reaction A +B <-> A’ + B’ A family of unknown function with experimental evidences with one available structure 17 substrats new reactions 2 Selection & Screening +Enzymatic screening Statistical analysis + Family partitioning Potential metabolites Set of sequences BLAST PDB Homology Modeling - MODELLER 3D Models Cavity Detection - FPOCKET 3D-Active Sites Structural Alignment - MULTALIGN Hierarchical Clustering - WEKA Specificity Determining Residues are determined by a log-likelihood analysis Pfam unknown family (DUF 846) Next Generation Sequencing technology has dramatically increased the number of available sequences in public databases. At the same time, many enzymatic activities (~22%) are orphans of protein sequence (Sorokina et al., 2014). The large amount of available protein sequences is an opportunity to discover enzymes associated to new reactions. We present here an integrated bioinformatics approach to reduce this lack of knowledge in metabolism and to propose new activity/protein associations for experimental validation. With this objective, the “New Enzymatic Activity” group of the LABGeM team is developing several methods. The CanOE method combines genomic and metabolic contexts to predict candidate genes for orphan enzymes (Smith et al., 2012). Currently, this approach is extended to the detection of conserved chemical transformation motifs in the metabolism (Sorokina et al., submitted). From a structural point of view, the ASMC (Active Site Modeling and Clustering) method finds and compares active site pockets to classify enzymes of a family and detects important residues for substrate specificity (de Melo-Minardi et al., 2010). These methods were successful applied to elucidate the enzymatic diversity of a protein family of unknown function (Bastard et al., 2014). Their results, associated with present knowledge, must be unified in a database allowing the elaboration of strategies for the selection of enzymatic families of interest. This work is supported by genomic and metabolic network data from MicroScope, a platform for microbial genome analyses (Vallenet et al., 2013). Exploration of archaeal enzyme activities: ARCHAEOACTOME research project Literature references Bastard, K. et al. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 10, 42–9 (2014). de Melo-Minardi, R. C., Bastard, K. & Artiguenave, F. Identification of subfamily- specific sites based on active sites modeling and clustering. Bioinformatics 26, 3075–82 (2010). Smith, A. A. T., Belda, E., Viari, A., Medigue, C. & Vallenet, D. The CanOE strategy: Integrating genomic and metabolic contexts across multiple prokaryote genomes to find candidate genes for orphan enzymes. PLoS Comput. Biol. 8, (2012). Sorokina, M., Stam, M., Médigue, C., Lespinet, O. & Vallenet, D. Profiling the orphan enzymes. Biol. Direct 9, 10 (2014). Vallenet, D. et al. MicroScope--an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 41, D636– 47 (2013). Sorokina et al. A novel metabolic network representation for the discovery of conserved modules of chemical transformations. Submitted. Mercier, J., Vallenet, D. GROOLS: Reactive Graph Reasoning for Genome Annotation. RuleML 2015 Conference Active Sites Classification (ASMC) The CanOE strategy Reaction Molecular Signature Network The dynamics of enzyme discoveryMicroScope From genomes to biological systems Reactions sharing a same RMS Reaction Network reduction into a RMS Network Microbial genome analysis Metabolic network >3,900 genomes 1-10 Mb ASMC method Classification of a family into groups of similar active sites NEA team Workbench Data Integration Orphan Enzymes Grools Structural Analysis Metabolic Network MicroScope