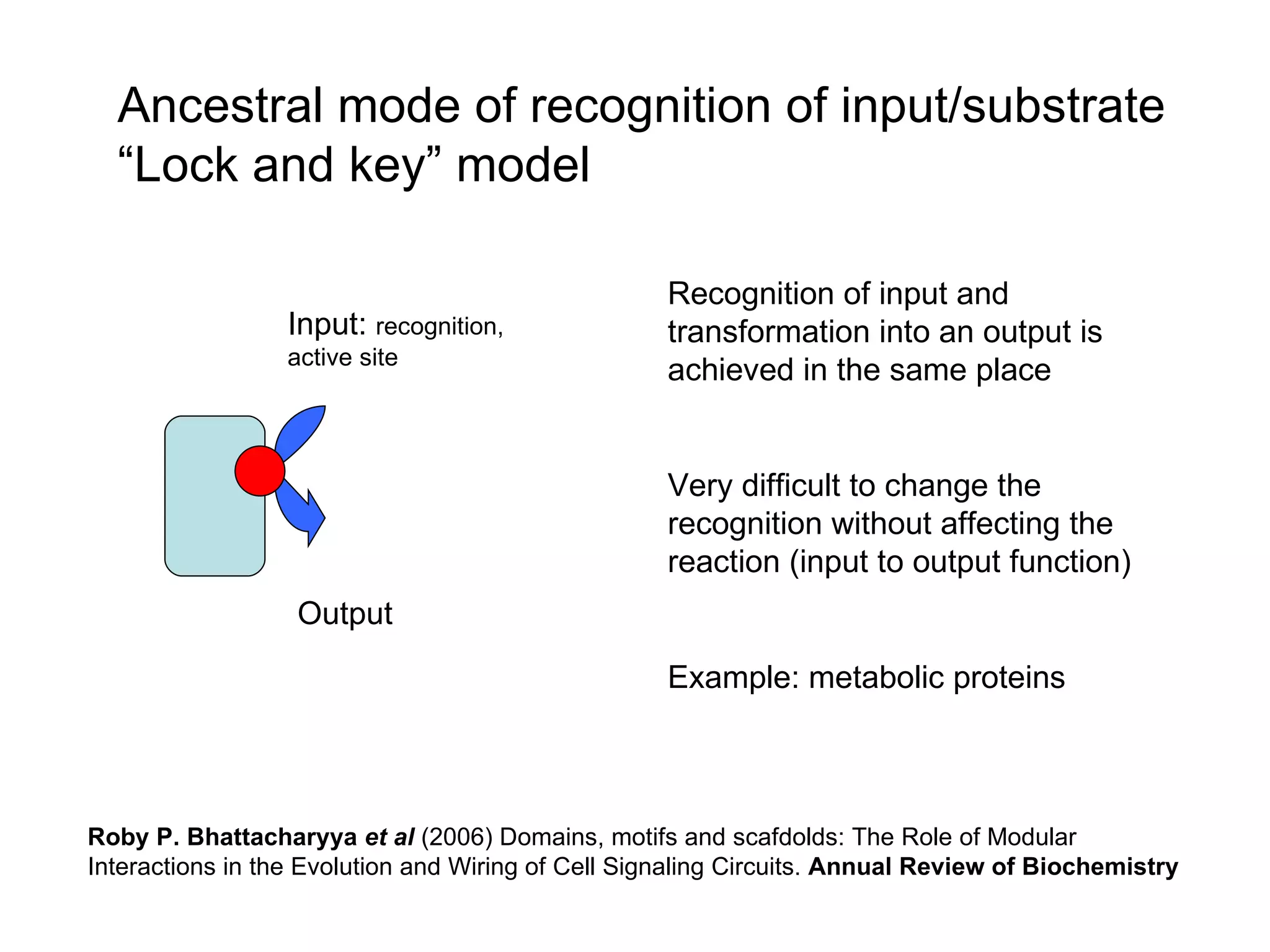
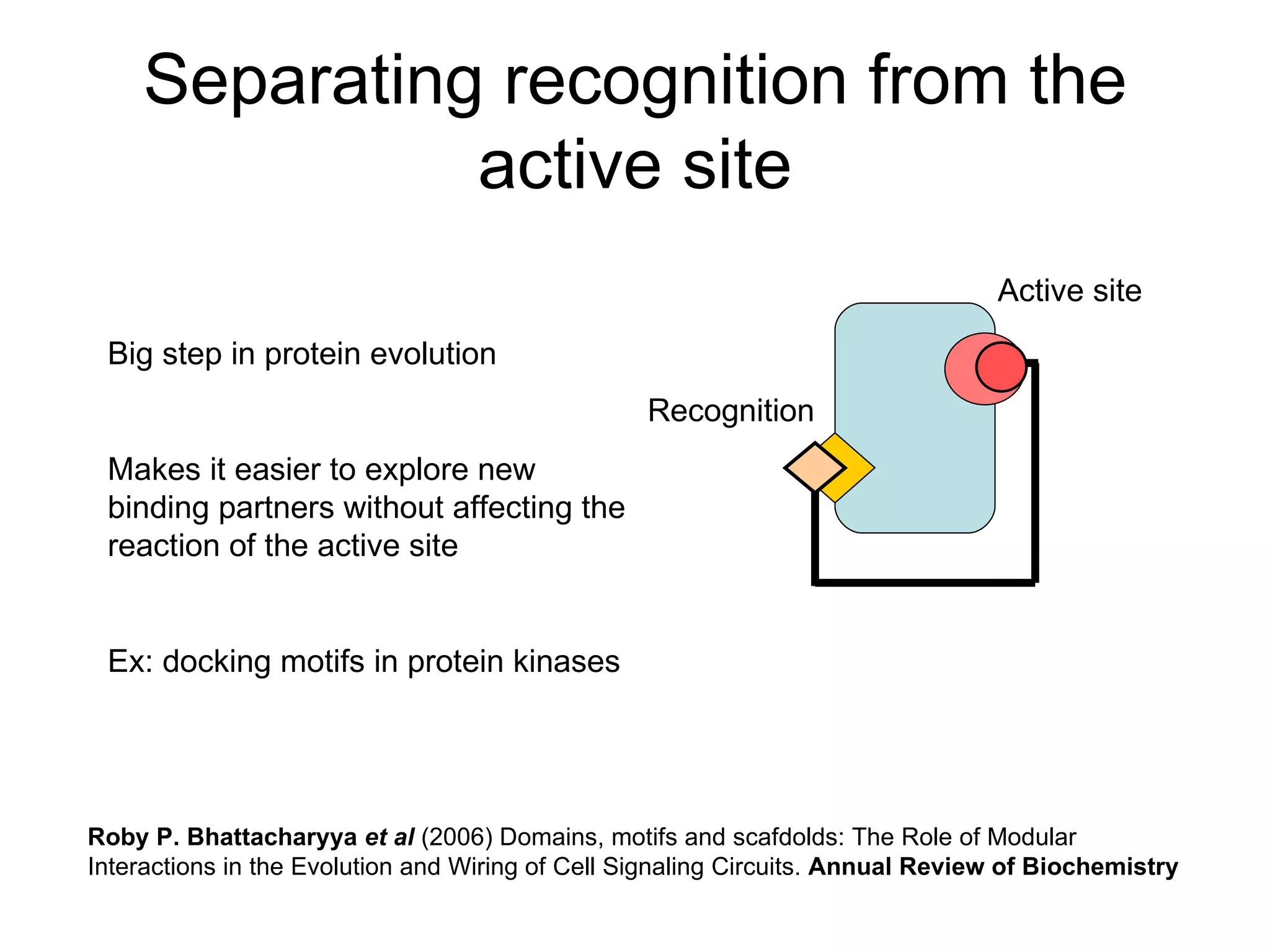
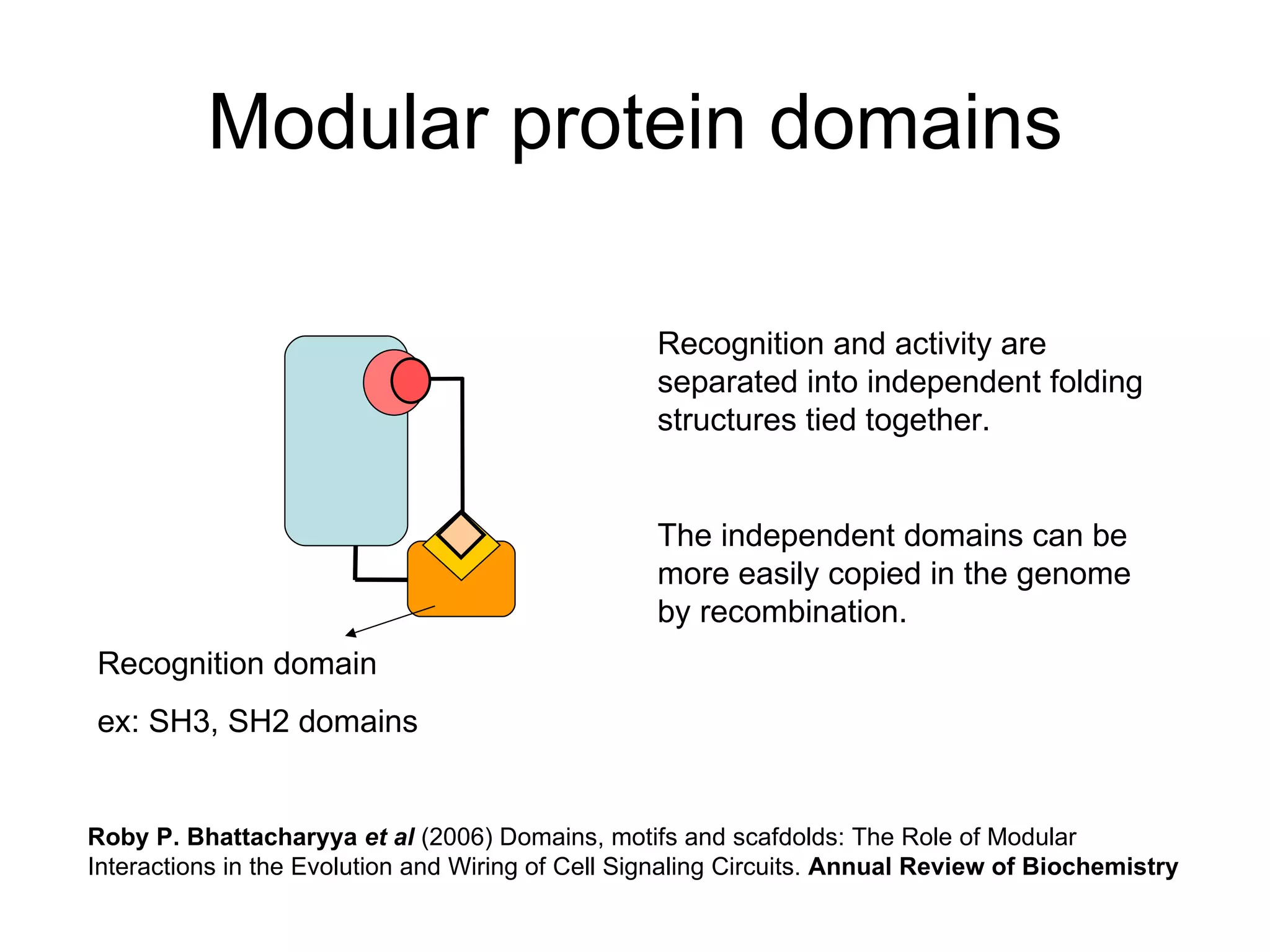
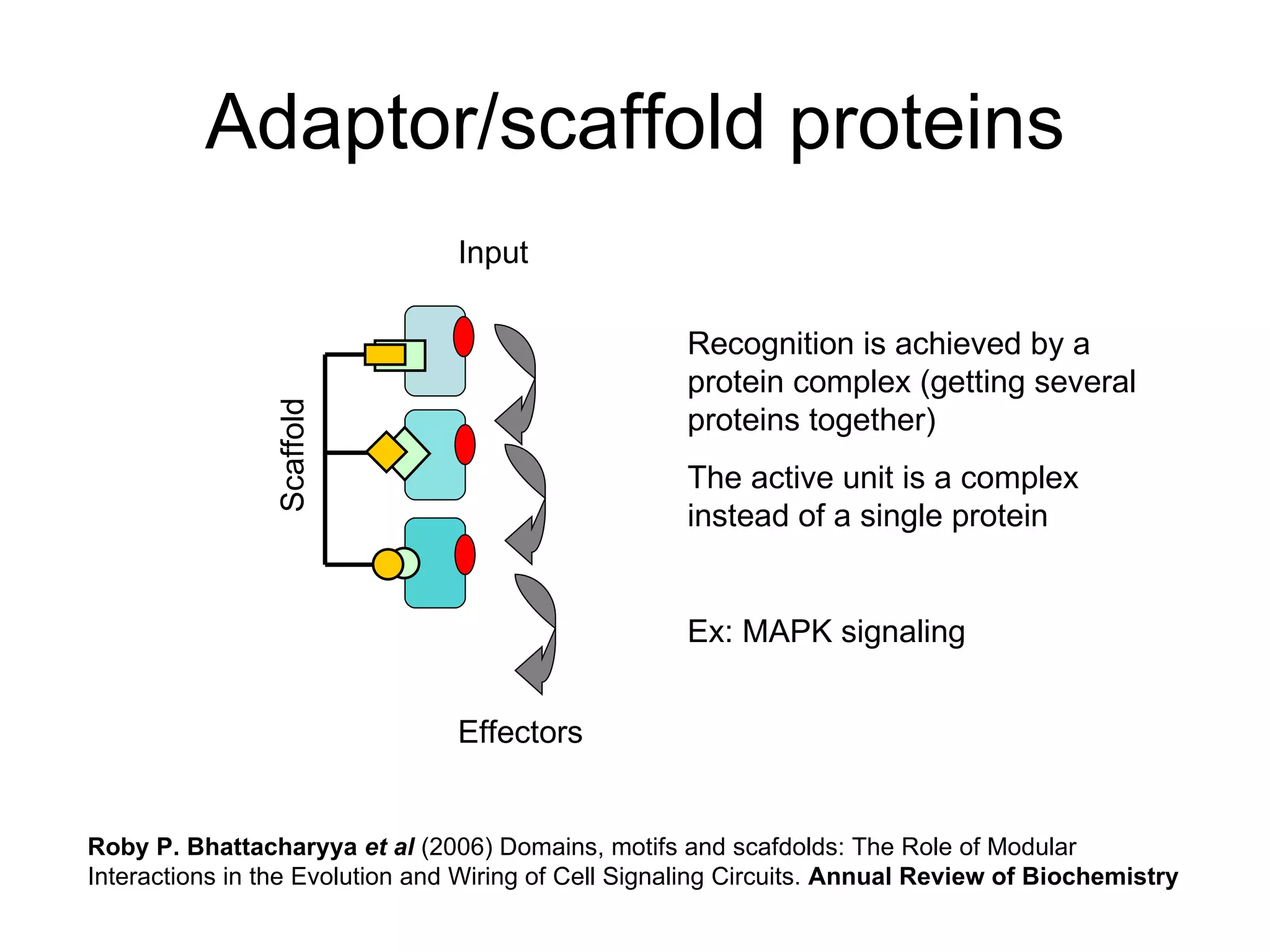
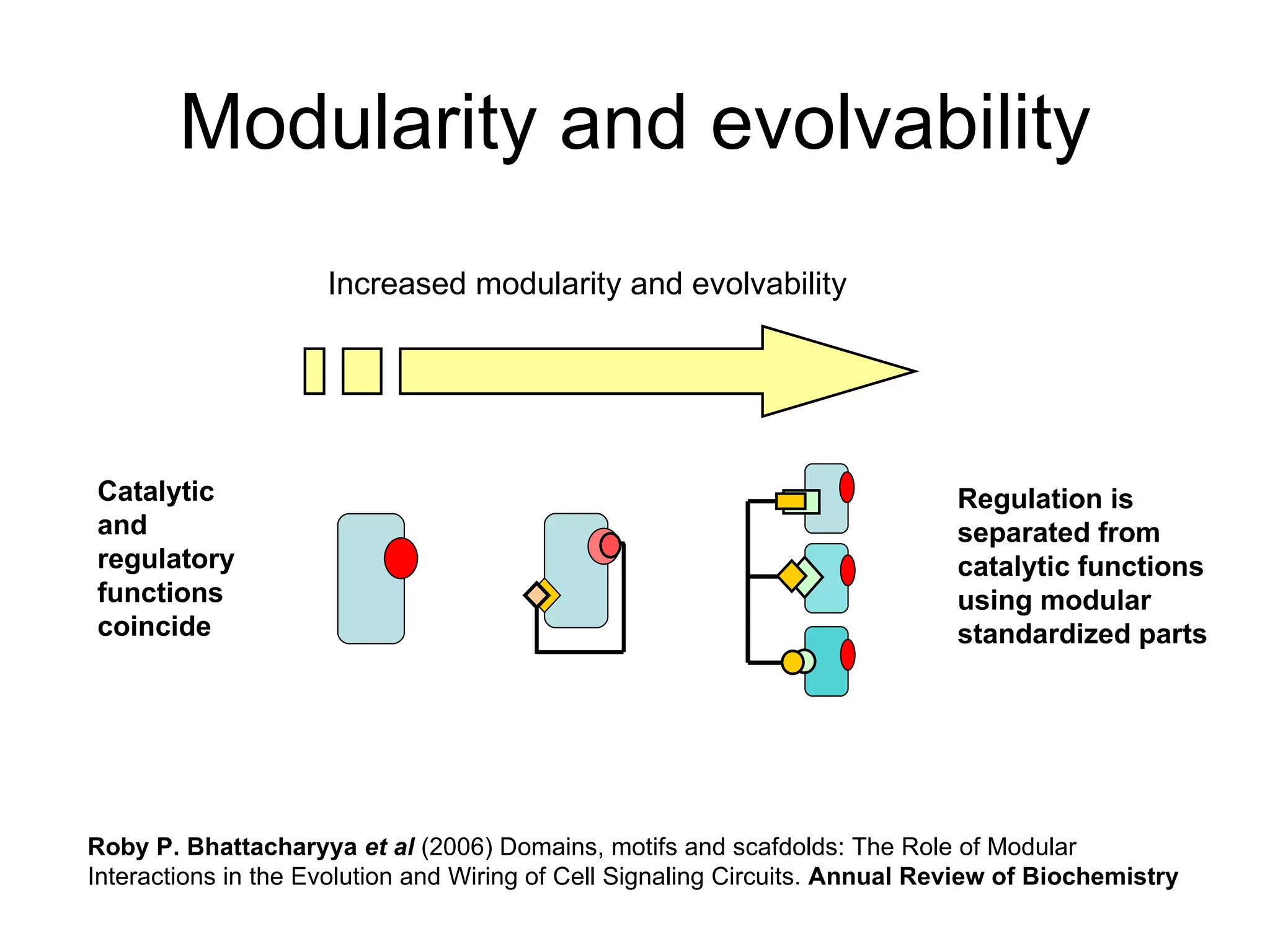
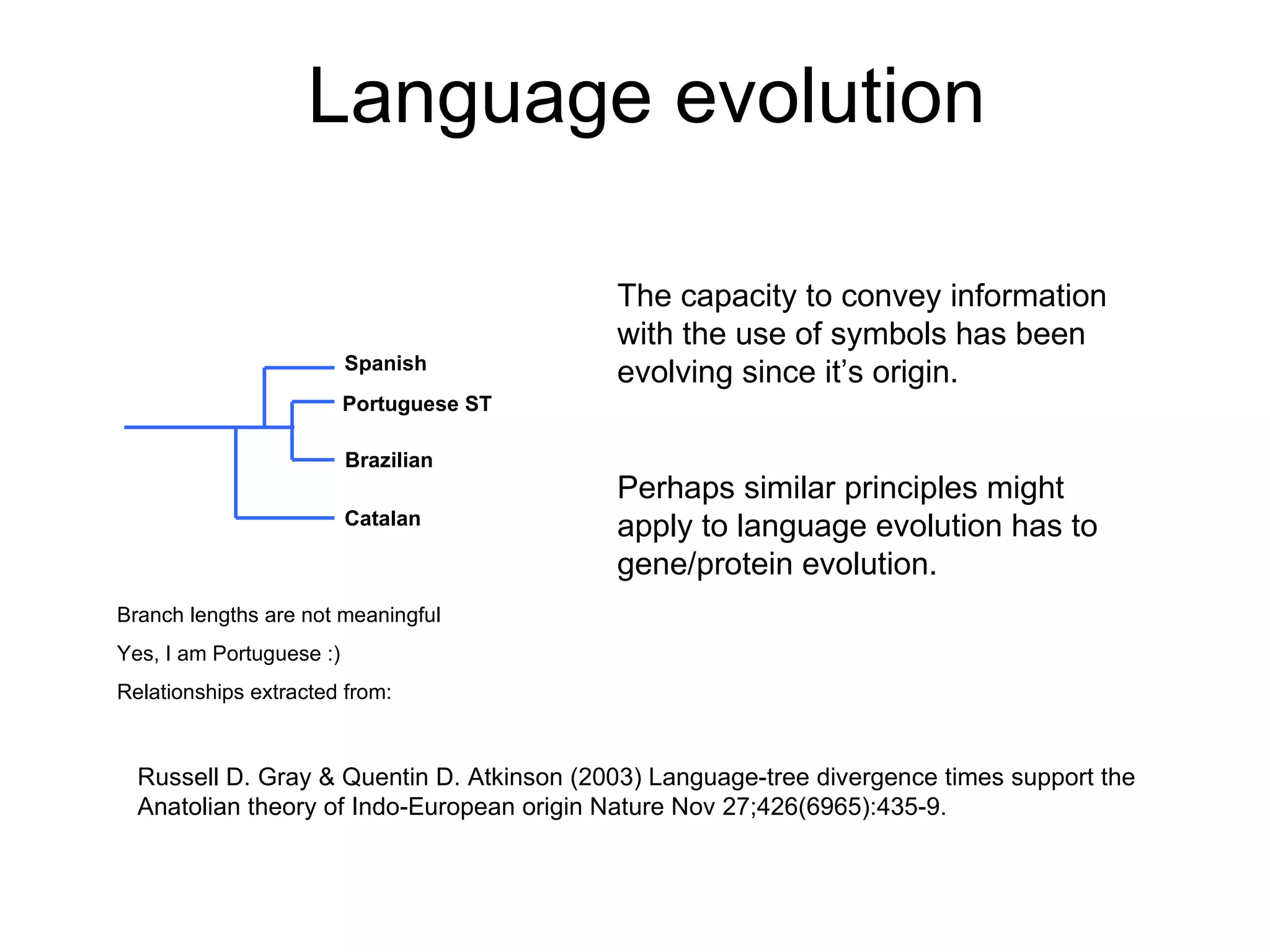
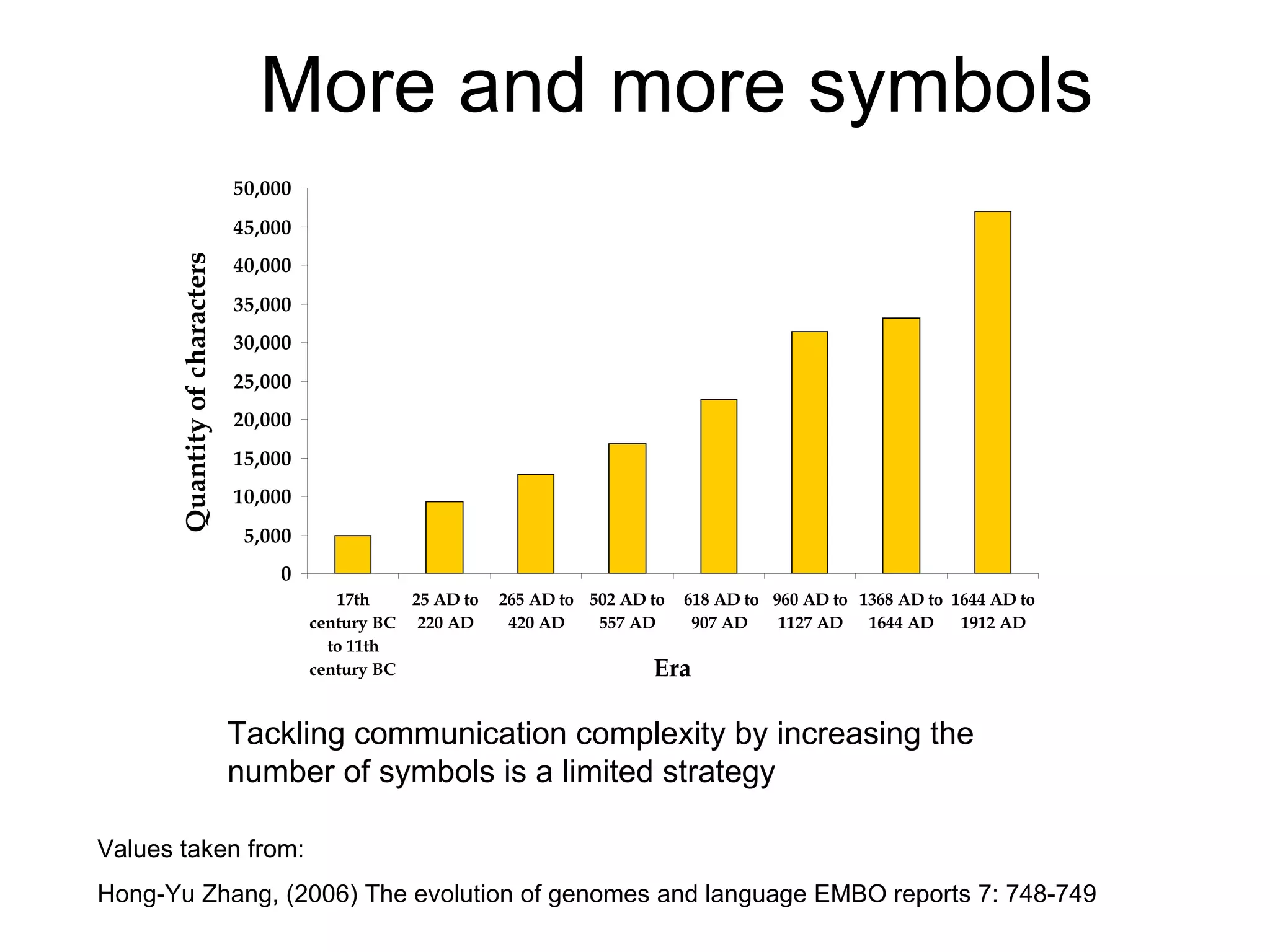
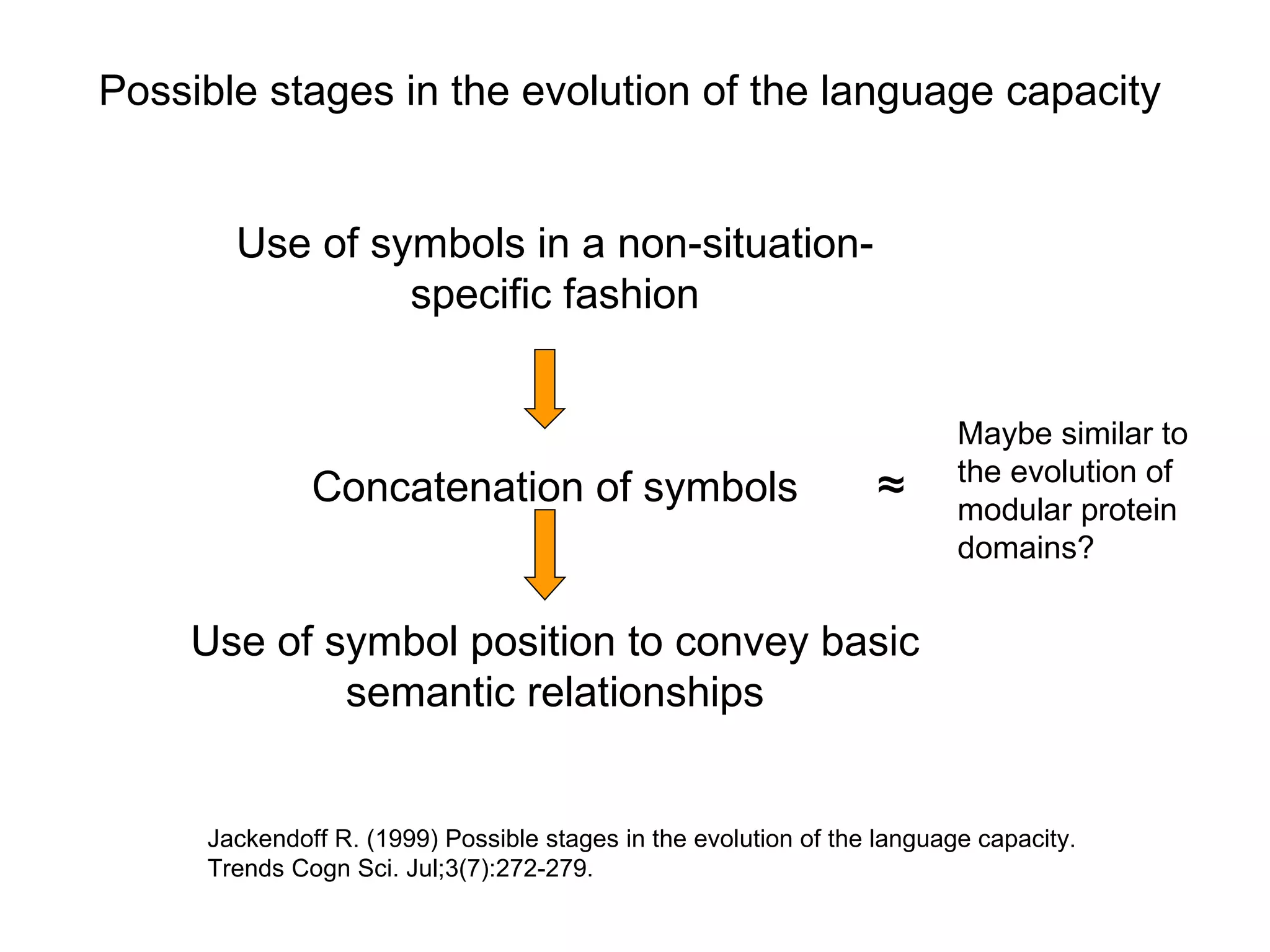
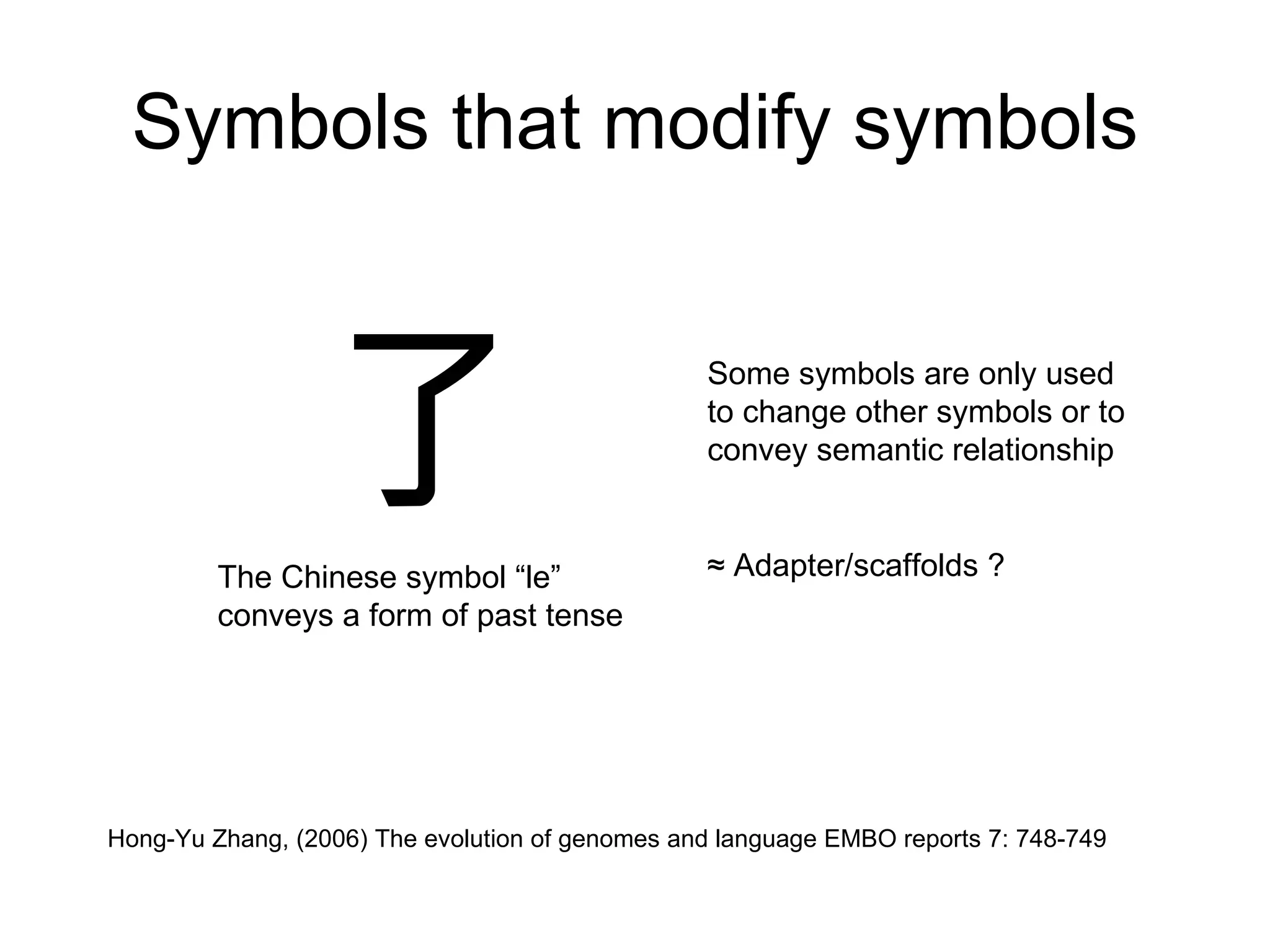
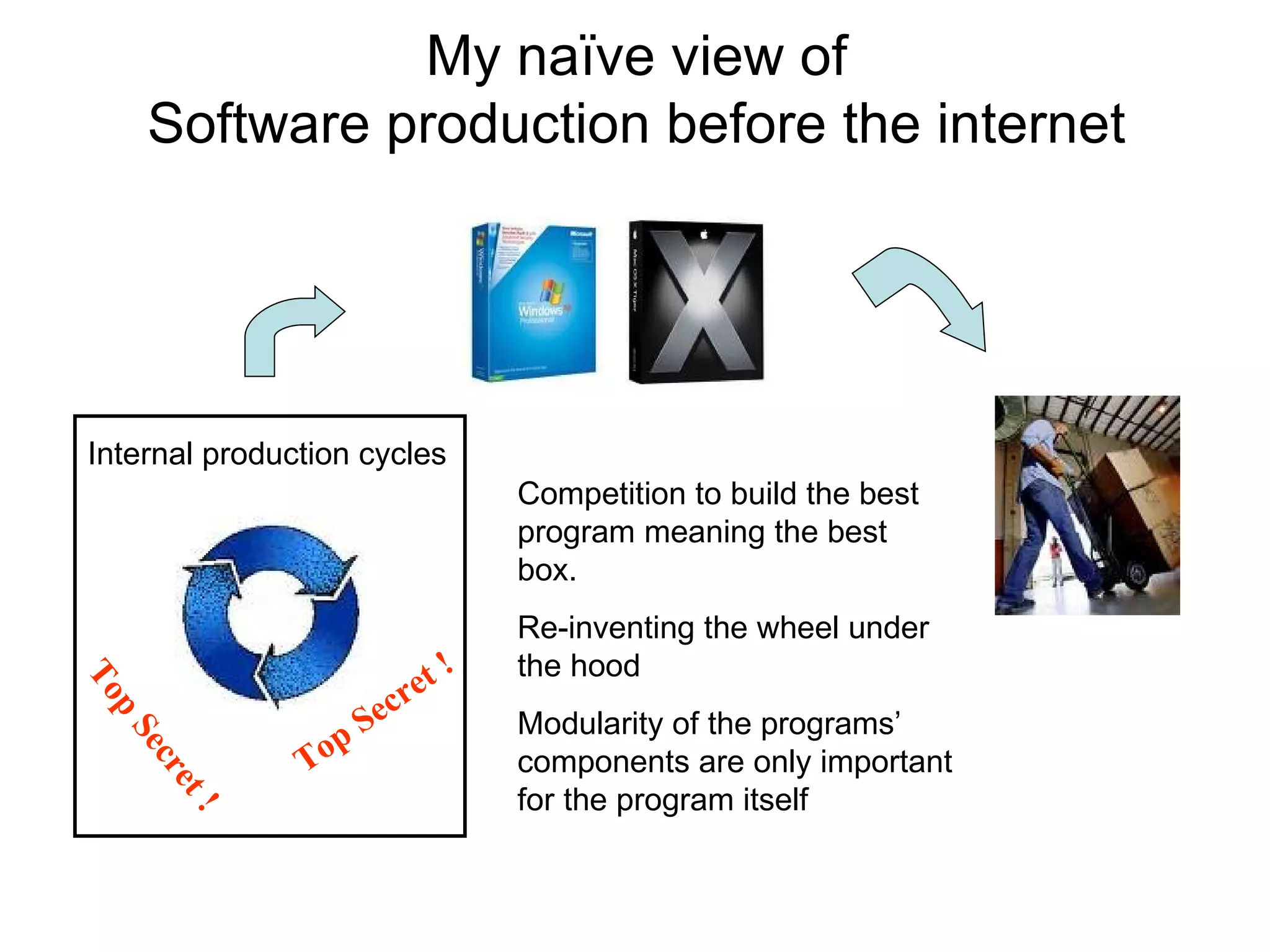
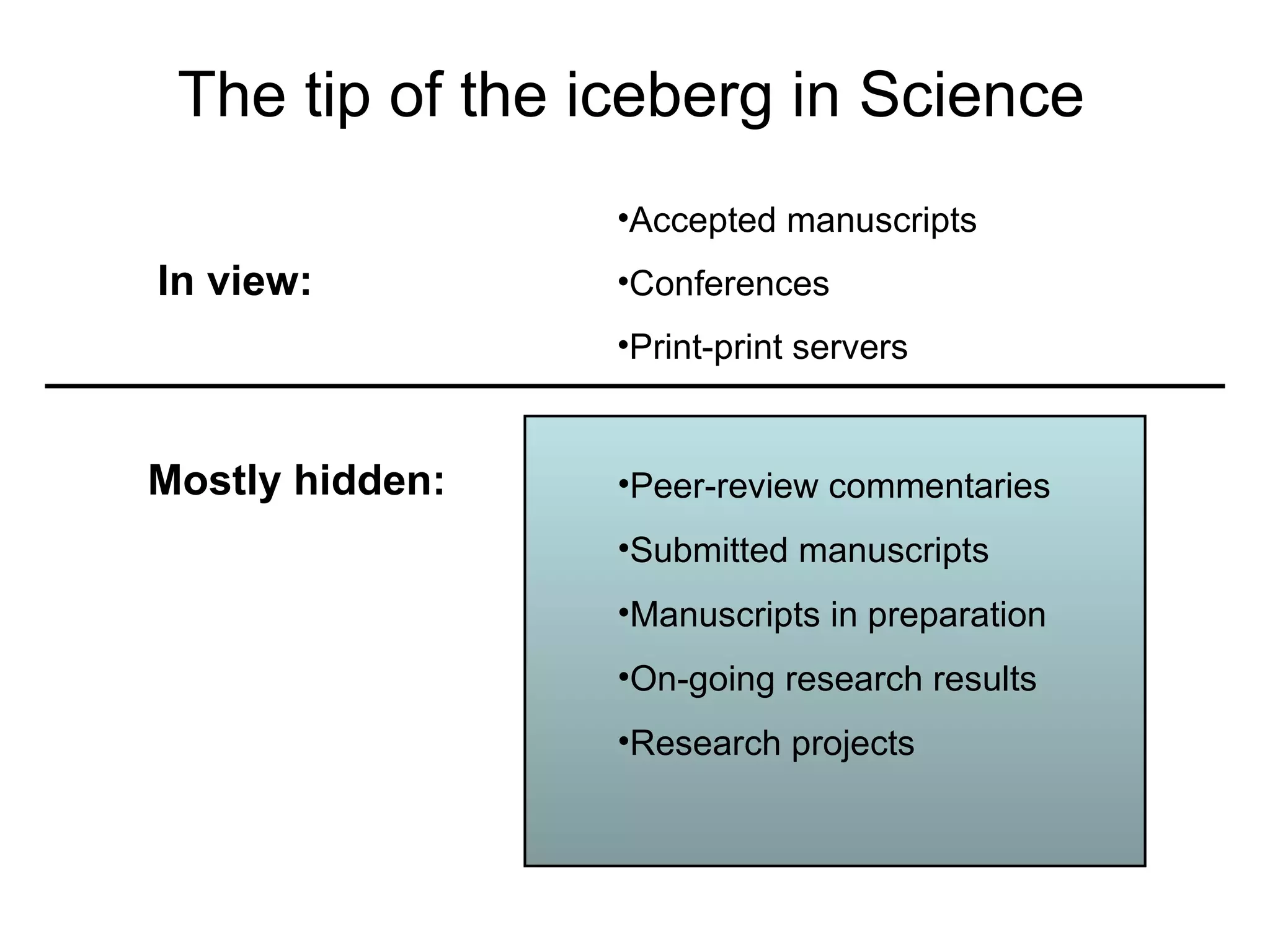
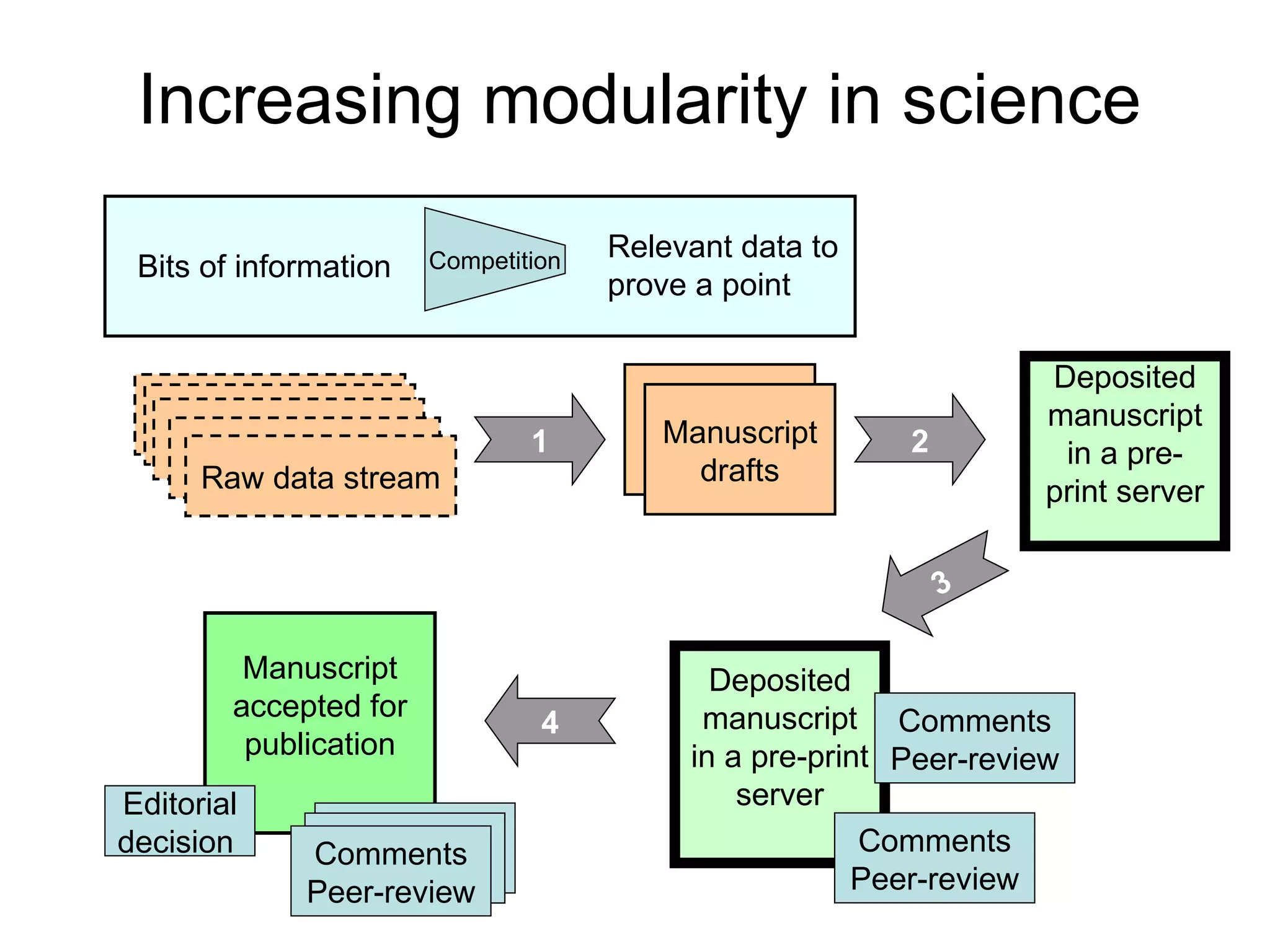
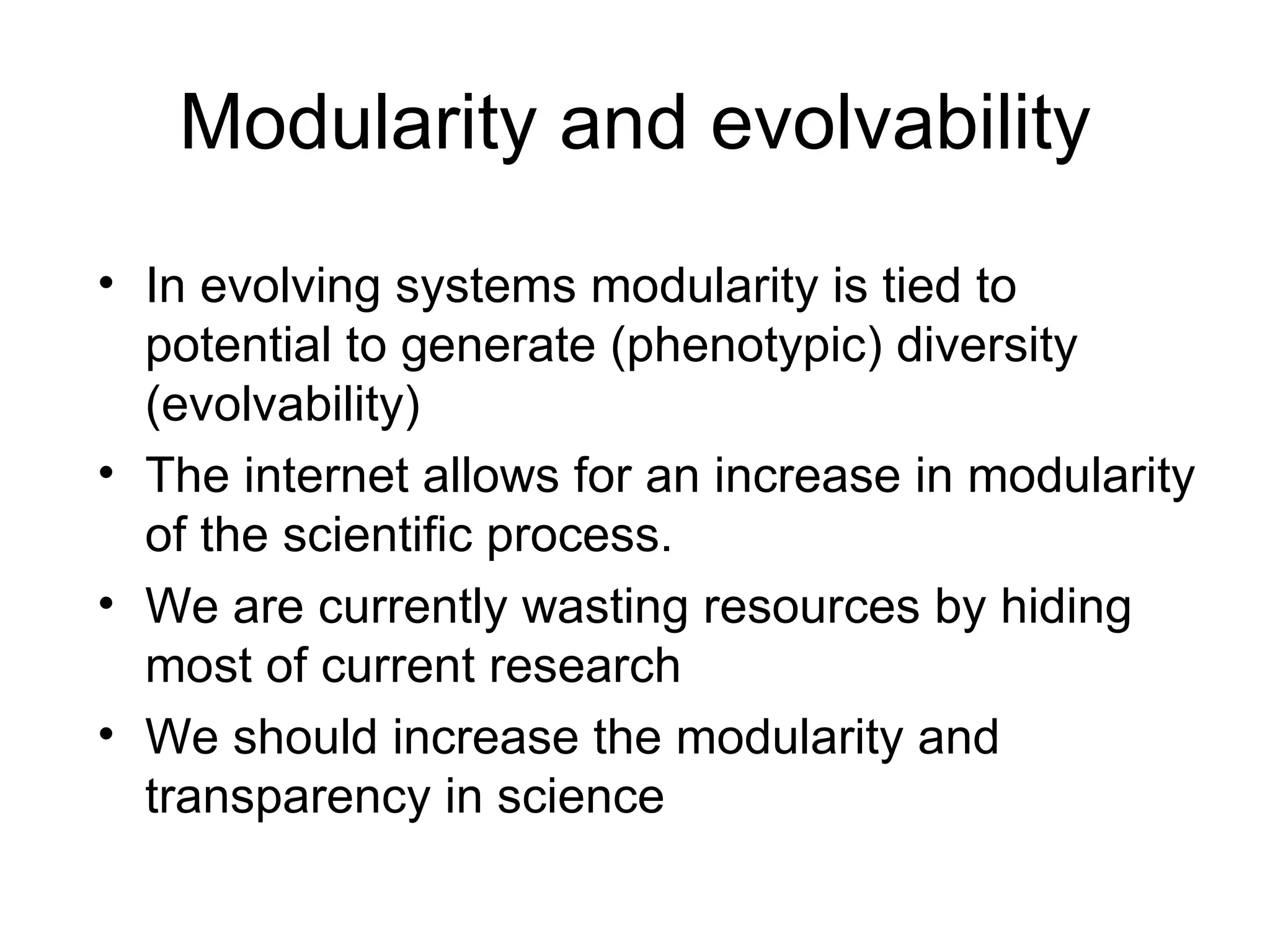
The document discusses how modularity has allowed for increased evolvability in biological and technological systems by separating functions and allowing independent development of modules. It argues that a similar increase in modularity could benefit scientific communication by making more research processes and results transparent earlier through preprint servers, comments, and finer-grained publications. This would reduce wasted effort and increase opportunities for collaboration.